Abstract
Performance characteristics of a hepatitis C virus (HCV) RNA quantification assay comprised automated specimen extraction [COBAS AmpliPrep (CAP) using total nucleic acid isolation reagents (TNAI)], and real-time polymerase chain reaction [COBAS TaqMan 48 HCV with analyte-specific reagents (CTM48)] were determined. CAP TNAI/CTM48 performed linearly from approximately 2.0 to at least 6.7 log10 IU/ml for HCV genotypes (Gts) 1, 2, and 3. The limit of detection for the World Health Organization International Standard was 23 IU/ml. Variabilities ranged from 1.3 to 2.1%. Excellent quantitative agreement was observed in clinical samples using CTM48 and two different methods for HCV RNA extraction (CAP TNAI and BioRobot M48; regression line slope, 0.98; y-intercept, 0.11; R2, 0.98; mean difference, 0.003). Good agreement was also observed between CAP TNAI/CTM48 and COBAS Amplicor Monitor (regression line slope, 0.94; y-intercept, 0.08; R2, 0.96), although HCV RNA concentrations were on average greater by COBAS Amplicor Monitor (mean difference −0.27 log10 IU/ml). Better overall agreement was observed for Gt 1 than non-Gt 1 specimens when comparing extraction and quantification methods; however, no consistent genotype-dependent quantification bias was observed. These data suggest that CAP TNAI/CTM48 offers an alternative method for the quantification of HCV in plasma samples.
Hepatitis C virus (HCV) molecular tests are currently the standard of care for diagnosis of acute and chronic HCV infections and monitoring for therapeutic efficacy. Tests that demonstrate a low limit of detection (LOD) are essential for both diagnosis and assessment of HCV clearance after treatment. A broad dynamic range is also important since HCV viral loads before antiviral therapy often exceed 6.0 log10 IU/ml. Assay performance is dependent on multiple factors, including nucleic acid extraction, amplification, and detection methods.
Automated nucleic acid extraction platforms differ in their robotics, chemistries, input/elution volumes, and sample throughput. Some use target-specific reagents, whereas others isolate DNA, RNA, or both. A major advantage of total nucleic acid isolation reagents (TNAI) is that DNA and RNA targets can be detected from a single extraction.
A variety of automated extraction platforms have been used to isolate HCV RNA from clinical samples. Evaluations using the BioRobot M48 and the BioRobot 9604 (Qiagen, Germantown, MD), the m1000 (Abbott Laboratories, Abbott Park, IL), the NucliSens Extractor (bioMerieux, Durham, NC), the MagNA pure LC, and AmpliPrep (Roche Diagnostics, Indianapolis, IN) have been reported.1,2,3,4,5,6,7 These studies demonstrate the reproducibility, sensitivity, and enhanced throughput of these instruments.
We have investigated the performance of the COBAS AmpliPrep (CAP)-automated extraction instrument using the TNAI kit followed by real-time amplification and detection with COBAS TaqMan 48 (CTM48). The CAP TNAI reagents purify RNA and DNA from serum or plasma on the CAP instrument. CTM48 utilizes TaqMan chemistry to amplify, detect, and quantify sequences in the 5′-untranslated region of HCV. The goal of this study was i) to determine analytical performance parameters, including measurable range, LOD, and precision of HCV Gt 1, 2, and 3 quantification by CAP TNAI/CTM48; ii) to correlate quantitative data generated by CTM48 using two different automated extraction methods (CAP TNAI versus BioRobot M48); iii) to compare quantification between CAP TNAI/CTM48 and COBAS Amplicor Monitor (CAM); and iv) to determine whether quantification by CAP TNAI/CTM48 was susceptible to genotype-dependent bias through a comparative analysis of Gt 1 and non-Gt 1 data.
Materials and Methods
Viruses for Analytical Studies
Six preparations of a Gt 1 virus were used for assay calibration (6.7, 5.7, 4.7, 3.7, 2.7, and 1.7 log10 IU/ml; AcroMetrix, Benicia, CA). Analytical verification studies (measurable range, LOD, and precision) were performed with a commercial panel of clinical specimens containing HCV Gt 1a, Gt 2b, and Gt 3a viruses (HemaCare BioSciences, Inc., Fort Lauderdale, FL). HCV Gt 1a and Gt 2b panel members were quantified by the manufacturer using HCV CTM48 analyte-specific reagent; HCV Gt 3a was quantified by CAM, version 2.0 (Roche Diagnostics). LOD was also determined using the World Health Organization (WHO) Second International Standard for Hepatitis C Virus RNA for Genomic Amplification Technology Assays (NIBSC code: 96/798; National Institute for Biological Standards and Control, Hertfordshire, UK). Viruses used to evaluate assay performance were requantified by CTM48 HCV analyte-specific reagent (Roche Diagnostics) using BioRobot M48 (Qiagen) extraction to verify the stated concentrations. New values were assigned if the concentration observed after requantification differed from the expected value by a factor of 1.5-fold or greater. HCV Gt 1, Gt 2, and Gt 3 panel members were confirmed to contain 6.8, 6.7, and 6.8 log10 IU/ml. Viruses were diluted in EDTA human plasma (BBI Diagnostics, West Bridgewater, MA) for measurable range, LOD, and precision experiments. The plasma contained no detectable anti-HCV antibodies and HCV RNA as confirmed by the manufacturer.
RNA Extraction Methods
HCV RNA for performance verification was extracted by the CAP instrument using TNAI reagents (Roche Diagnostics). The TNAI Kit process consists of the following five steps: 1) digestion by protease to facilitate the release of RNA and DNA; 2) protein denaturation and nucleic acid solubilization/stabilization by proprietary lysis reagent; 3) nucleic acid binding to the silica surface of the added magnetic glass particles due to chaotropic salts and the high ionic strength of the lysis reagent; 4) reduction of salt concentration and removal of unbound substances and impurities, such as denatured proteins, cellular debris, and potential polymerase chain reaction (PCR) inhibitors such as hemoglobin by washing; and 5) elution of purified nucleic acids at elevated temperatures. In the protocol used for this study, plasma input volume was 650 μl (500 μl processed), and the elution volume was 75 μl. Residual, unprocessed plasma (approximately 150 μl) remained in the sample input tube. Quantification standard (286 μl) was added to quantification standard diluent (3184 μl), resulting in a final concentration of 22 copies/μl after extraction.
Plasma samples for correlation studies were also extracted on the M48 using the Infectious Diseases, Virus Mini software protocol (Qiagen) and the MagAttract Virus Mini Kit (Qiagen). Quantification standard reagent was added to the kit lysis buffer, resulting in a final concentration of 30 copies/μl. The sample input volume was 400 μl, and the elution volume was 125 μl. For quantification by CAM, samples (200 μl) were extracted manually with the manufacturer's guanidinium-based sample preparation method using proprietary reagents. Extracted RNA was resuspended in 400 μl of proprietary specimen diluent as per the manufacturer's recommendation.
HCV RNA Amplification and Detection
For CTM48, master mix was activated by combining master mix (1.4 ml) with manganese (170 μl). The activated master mix (50 μl) was added to each K-tube (Roche Diagnostics) in the K-Carrier (Roche Diagnostics) followed by 50 μl of test sample. The K-Carrier was placed in the CTM48 instrument for reverse transcription, amplification, and detection. CAM was performed according to the manufacturer's instructions.
Design of Analytical Performance and Correlation Experiments
Measurable Range
The measurable range of CAP TNAI/CTM48 quantification was determined by testing nine concentrations of genotype 1, 2, and 3 viruses beginning with the neat samples (10 replicates per concentration in two runs of five replicates each). The criteria of linearity and acceptable variability (SD, ≤0.4, slightly more conservative than 0.5 log10, the acknowledged variability of PCR quantification) were used to define the upper and lower limits of quantification.
LOD
For LOD determinations, viruses were diluted to 100 IU/ml (in tenfold increments) and than further diluted to 40, 30, 20, 15, 10, and 7.5 IU/ml in EDTA human plasma. Twenty replicates were tested at each concentration, with the exception of the WHO standard at 15 and 10 IU/ml. The LOD was defined as the concentration at which 95% of replicates was detected as estimated by PROBIT.
Precision
CAP TNAI/CTM48 precision was assessed using Gts 1, 2, and 3 at 3.0 and 5.0 log10 IU/ml. For intrarun precision, 20 replicates were tested at each concentration. To calculate inter-run precision, 10 additional replicates were tested on two additional runs performed on two separate days, and the data were combined with the initial run of 20 replicates. The percentage CV was calculated from log-transformed viral load values.
Correlation of Quantification between CAP TNAI/CTM48, M48/CTM48, and CAM
To assess whether quantification by CAP TNAI/CTM48 and the verified in-house clinical M48/CTM48 correlated, 121 plasma specimens were tested by both methods (distribution by genotype is depicted in the legends of Figures 2 through 4). Samples were not tested simultaneously on both platforms but were first tested by the in-house clinical assay and then frozen at −70°C until CAP TNAI extraction and CTM48 testing. Specimens throughout the range of quantification were chosen for analysis. Data were logarithmically transformed before analysis. Genotypes were determined by direct sequencing of core-E1 sequences8 or VERSANT HCV genotyping assay (LiPA 2.0; Siemens Medical Diagnostics Solutions, Tarrytown, NY). Genotype could not be determined for six specimens. CAM was performed on a subset of these specimens that had HCV RNA concentrations greater than the lower quantification limit of this assay (600 IU/ml). Samples with HCV RNA concentrations greater than the upper quantification limit of CAM were diluted in normal human plasma before testing. Genotype-dependent bias in quantification was examined through a separate analysis of data from Gt 1 and non-Gt 1 specimens. All studies were performed following guidelines for human subject experimentation. Approval was received from the Johns Hopkins Medicine Institutional Review Board to conduct these experiments.
Figure 2.
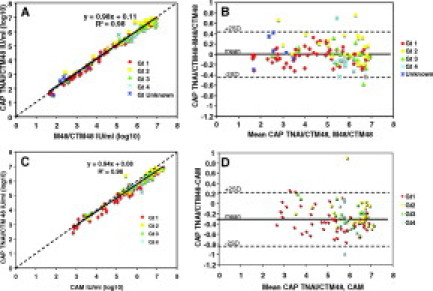
Quantitative agreement between two different extraction methods and two different quantitative methods: combined analysis of all samples. A: Quantitative correlation between CAP TNAI/CTM48 and M48/CTM48 (Gt 1, n = 61; Gt 2, n = 27; Gt 3, n = 16; Gt 4, n = 11; Gt unknown, n = 6). B: Difference in quantification between CAP TNAI/CTM48 and M48/CTM48. Mean, 0.00; SD, 0.22 log10 IU/ml. C: Quantitative correlation between CAP TNAI/CTM48 and CAM (Gt 1, n = 49; Gt 2, n = 23, Gt 3, n = 15; Gt 4, n = 11). D: Difference in quantification between CAP TNAI/CTM48 and CAM. Mean, −0.27 log10 IU/ml; SD, 0.26. Dashed line in A and C indicates theoretical trend line of complete agreement.
Figure 3.
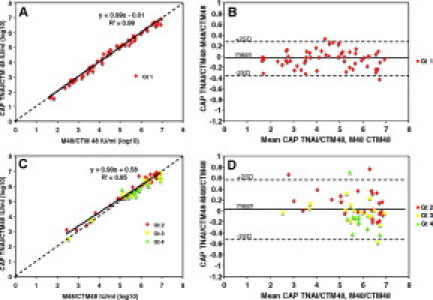
Quantitative agreement between two different extraction methods: separate analyses of Gt 1 and non-Gt 1 samples. A: Quantitative correlation between CAP TNAI/CTM48 and M48/CTM48 for Gt 1 samples (n = 61). B: Difference in quantification between CAP TNAI/CTM48 and M48/CTM48 for Gt 1 samples. Mean, −0.04; SD, 0.16. C: Quantitative correlation between CAP TNAI/CTM48 and M48/CTM48 for Gt non-1 samples (Gt 2, n = 27; Gt 3, n = 16; Gt 4, n = 11). D: Difference in quantification between CAP TNAI/CTM48 and M48/CTM48 for non-Gt 1 samples. Mean, 0.03; SD, 0.27. Dashed line in A and C indicates theoretical trend line of complete agreement.
Figure 4.
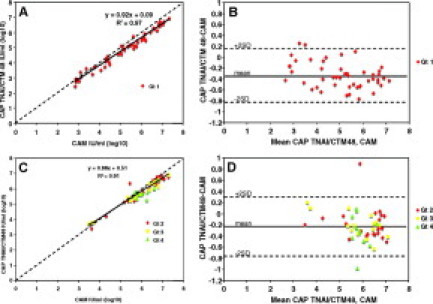
Quantitative agreement between two different quantification methods: separate analyses of Gt 1 and non-Gt 1 samples. A: Quantitative correlation between CAP TNAI/CTM48 and CAM for Gt 1 samples (n = 49). B: Difference in quantification between CAP TNAI/CTM48 and CAM for Gt 1 samples. Mean, −0.33; SD, 0.24. C: Quantitative correlation between CAP TNAI/CTM48 and CAM for Gt non-1 samples (Gt 2, n = 23; Gt 3, n = 15; Gt 4, n = 11). D: Difference in quantification between CAP TNAI/CTM48 and CAM for non-Gt 1 samples. Mean, −0.22; SD, 0.27. Dashed line in A and C indicates theoretical trend line of complete agreement.
Statistical Methods
Regression lines and their characteristics were determined using Microsoft Excel (Microsoft Office 2001; Microsoft Corp., Redmond, WA). LODs were determined by PROBIT regression analysis using nominal standard as an independent variable and number of positive results as the dichotomous variable.
Results
Measurable Range Using CAP TNAI/CTM48
The measurable range of CAP TNAI/CTM48 was determined for the three HCV genotypes (Gts 1, 2, and 3) that are most prevalent in our patient population. Measurable ranges (log10 IU/ml) were 2.4 to 6.8 for Gt 1 (Figure 1A), 1.8 to 6.7 for Gt 2 (Figure 1B), and 1.9 to 6.8 for Gt 3 (Figure 1C).
Figure 1.
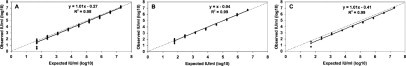
Measurable range (IU/ml) of different HCV genotypes by CAP TNAI/CTM48. A: Gt 1. B: Gt 2. C: Gt 3. Dashed line indicates theoretical trend line of complete agreement between expected and observed values.
LODs for HCV Gts 1, 2, and 3 and the WHO Standard Using CAP TNAI/CTM48
For the WHO Standard, all replicates were detected at 30 IU/ml, and 90% of replicates was detected at 20 IU/ml (Table 1), suggesting that the LOD was between 20 and 30 IU/ml (23 IU/ml by PROBIT). For Gt 1, 100% of replicates was detected at 15 IU/ml, with 90% of replicates detected at 10 IU/ml, suggesting that the LOD was between 15 and 10 IU/ml (11 IU/ml by PROBIT). For Gt 2, 100% of replicates was detected at 10 IU/ml, and 90% of replicates was detected at 7.5 IU/ml, suggesting that the LOD was between 7.5 and 10 IU/ml (8 IU/ml by PROBIT). For Gt 3, 100% of replicates was detected at 20 IU/ml, with 65% of replicates detected at 15 IU/ml, suggesting that the LOD was between 15 and 20 IU/ml (16 IU/ml by PROBIT).
Table 1.
LODs of AmpliPrep TNAI/CTM48 HCV
| % Replicates detected (no. positive/total no. of replicates) |
||||
|---|---|---|---|---|
| Concentration (IU/ml) | WHO standard | Gt 1 | Gt 2 | Gt 3 |
| 30 | 100 (20/20) | NDa | ND | ND |
| 20 | 90 (18/20) | ND | ND | 100 (20/20) |
| 15 | 15 (2/12) | 100 (20/20) | ND | 65 (13/20) |
| 10 | 10 (1/10) | 90 (18/20) | 100 (20/20) | ND |
| 7.5 | ND | 85 (17/20) | 90 (18/20) | ND |
ND, not done.
Precision of the CAP TNAI/CTM48
Intra- and inter-run precision was determined for the CAP TNAI/CTM48 method (Table 2). The data demonstrate that intra- and interassay precision was similar for all three genotypes. The greatest degree of variability was observed between assays of the low concentration replicates (3.0 log10 IU/ml; Table 2).
Table 2.
Precision of COBAS AmpliPrep TNAI/CTM48
| Genotype 1 |
Genotype 2 |
Genotype 3 |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Intra-run |
Inter-run |
Intra-run |
Inter-run |
Intra-run |
Inter-run |
|||||||
| 3.0 | 5.0 | 3.0 | 5.0 | 3.0 | 5.0 | 3.0 | 5.0 | 3.0 | 5.0 | 3.0 | 5.0 | |
| Mean | 3.1 | 4.8 | 3.1 | 4.8 | 3.1 | 5.3 | 3.1 | 5.3 | 3.1 | 4.8 | 3.1 | 4.8 |
| SD | 0.04 | 0.1 | 0.1 | 0.1 | 0.04 | 0.1 | 0.1 | 0.1 | 0.04 | 0.1 | 0.1 | 0.1 |
| %CV | 1.4 | 1.4 | 2.1 | 1.8 | 1.3 | 1.5 | 2.4 | 1.3 | 1.4 | 1.4 | 2.1 | 1.8 |
All values are log10 IU/ml.
CTM48 Quantification of Clinical Samples: Comparison between Two Different Automated Extraction Methods and Two Different Quantification Assays
A comparison of CTM48 quantification after extraction by CAP TNAI and M48 demonstrated excellent correlation, with regression line slope of 0.98 and y-intercept of 0.11 (Figure 2A). Bland Altman analysis3 further showed very little difference in quantification after extraction with two different methods as demonstrated by negligible mean difference (0.003) and a small degree of variability (SD of data was 0.22 log10 IU/ml or 1.6-fold, and most results fell within two standard deviations of the mean; Figure 2B).
The agreement between quantitative data obtained with two different assays (CAP TNAI/CTM 48 versus CAM) was not as good as that observed with a single assay using two different extraction methods, as demonstrated by the regression slope (0.94; Figure 2C). HCV RNA concentrations obtained with CAM were, on average, 0.27 log10 IU/ml greater than those obtained with CAP TNAI/CTM 48 (Figure 2C). However, the degree of quantitative variability in relation to the mean difference was small (SD 0.26; Figure 2D), similar to that observed when different extraction methods were compared.
To better demonstrate whether a genotype-dependent bias in quantification occurred with the CAP TNAI/CTM 48 assay, non-Gt 1 samples from the correlation studies above were analyzed separately from Gt 1 samples. For non-Gt 1 samples, HCV RNA concentrations obtained by CAP TNAI/CTM48 were linearly related to those obtained by comparator methods (Figure 3C, M48/CTM48, and Figure 4C, CAM). However, the agreement was not as good as that observed for Gt 1 samples [CAP TNAI/CTM48 versus M48/CTM48 Gt 1 regression line slope 0.99, y-intercept 0.01 (Figure 3A); non-Gt 1 regression line slope 0.90, y-intercept 0.58 (Figure 3C); CAP TNAI/CTM48 versus CAM Gt 1 regression line slope 0.92, y-intercept 0.09 (Figure 4A); and non-Gt 1 regression line slope 0.88, y-intercept 0.51 (Figure 4C)]. Bland Altman analysis indicated that the mean difference in quantification of Gt 1 and non-Gt 1 samples was similar when comparing CAP TNAI/CTM48 to M48/CTM48 [Gt 1, −0.04 (Figure 3B); non-1 Gt, 0.03 (Figure 3D)] or CAP TNAI/CTM48 to CAM [Gt 1, −0.33 (Figure 4B); non-1 Gt, −0.22 (Figure 4D)]. For Gt 1 samples, differences in quantification were distributed fairly evenly above and below the mean throughout the range of quantification, and the number of outliers (>2 SD) was low. For non-1 Gt samples, differences in quantification were evenly distributed above and below the mean at viral loads greater than 5.0 log10 IU/ml. At concentrations less than 5.0 log10 IU/ml, non-Gt 1 CAP TNAI CTM48 data were greater than the comparator assays (M48/CTM48 and CAM, Figures 3D and 4D) but within an acceptable range of 2 SD (approximately 0.5 log10 IU/ml), with the exception of a single sample. Finally, greater quantitative disagreement was observed sporadically in non-Gt 1 than in Gt 1 samples as indicated by the number of non-Gt 1 specimens with differences in quantification greater than two standard deviations from the mean (Figure 3, compare B to D) and by the magnitude of these differences (disagreement between CAP TNAI/CTM48 and CAM for two non-Gt 1 specimens approached 1.0 log10 IU/ml; Figure 4, compare B to D).
Discussion
Our data demonstrate that CAP TNAI/CTM48 has a measurable range of at least 5 orders of magnitude for genotypes 1, 2, and 3, similar to that described for assays using CTM48 HCV reagents and other extraction methods.2,4,9,10 The upper limit of quantification was approximately 7.0 log10 IU/ml. The lower limit of quantification was estimated to be 2.0 log10 IU/ml. Levels below this, particularly with Gt 1, showed too much variability to quantify accurately.
The LODs of the CAP TNAI/CTM48 assays were lower than the M48/CTM48 assay.2 This may be explained by the differences in the input and output sample volumes for the two extraction platforms. The M48 uses a total nucleic acid extraction kit (Virus Mini; Qiagen) with an input volume from 50 to 400 μl and an elution volume of 50 to 150 μl. We verified performance of this extraction platform coupled with the CTM48 assay with an input volume of 400 μl and an elution volume of 125 μl. Variable sample volumes (50, 100, 200, 500, or 800 μl) can be processed by the CAP instrument. However, the instrument requires input volumes from 250 to 1000 μl for accurate automated transfer to the sample processing unit. The elution volume is fixed at 75 μl. We verified the CAP TNAI coupled with the CTM48 assay with an input volume of 650 μl (500 μl processed). The concentration factors for CAP and M48 were 8.7 and 3.2, a difference of greater than twofold. The observed LOD was comparable to other real-time PCR assays that used RNA extracted from similar plasma volumes [10 IU/ml when extracted by MinElute (Qiagen) from 500 μl of plasma2,9] and slightly lower volumes than assays that used less plasma (70 to 200 IU/ml for 200 μl of plasma4,10). Of note, LODs for the WHO standard and nominal genotyped standards were similar although not identical, potentially due to a number of factors, including variability introduced during dilution and the use of different assays to quantify standards.
There was a single difference in the instrument setting for the CTM48 assay used with the CAP extraction platform compared to the CTM48 assay used with the M48 extraction platform. The reverse transcription temperature was 59°C for samples extracted on the M48 and 64°C for samples extracted by CAP. The increase in temperature was implemented to decrease possible genotype bias by eliminating secondary structure. This temperature change appeared to have little impact on quantification for samples tested by both methods.
Intra- and inter-run precision at the 3.0 and 5.0 log10 IU/ml concentrations for Gts 1, 2, and 3 was within an acceptable range for an automated extraction platform and a real-time PCR method. Variability was comparable for the two concentrations tested; however, variability may be greater at concentrations lower than 3.0 log10 IU/ml.
Genotype bias has been observed for HCV RNA quantification assays.11,12,13 We did not observe major genotype bias with the commercial Gt 2 and Gt 3 panel members or with the clinical specimens from patients infected with Gt 2, Gt 3, and Gt 4 viruses. Although the level of agreement was not as good as Gt 1 specimens and sporadic disagreement of up to one log was observed, the overall performance of CAP TNAI/CTM48 on non-Gt 1 specimens was acceptable, given that mean differences in quantification were comparable to Gt 1 specimens and that quantitative differences exhibited satisfactory standards of deviation (approximately 0.5 log, the accepted degree of quantitative variability for PCR).
Of note, a larger variation in quantification between CAP TNAI/CTM48 and CAM for non-Gt 1 samples (mean difference of −0.77 ± 0.40) has been reported.14 The TNAI extraction in this previous study was based on magnetic silica generic nucleic acid purification, similar to our method; however, CAP-processing steps are slightly different. The potential impact of these changes on genotype-dependent quantification bias is unknown.
Our study was designed to determine the level of agreement between two different extraction methods and two different quantitative assays for the most common genotypes in our patient population throughout the range of quantification. We were able to meet this design specification for the Gt 1 sample cohort; however, our non-1 Gt analysis is somewhat constrained by a paucity of specimens with HCV RNA concentrations less than 5.0 log10 IU/ml. We routinely store specimens for 2 years before discarding. Unfortunately, we were unable to expand the number of non-Gt 1 samples in this range despite a 2-year look-back.
One major technical advantage of the CAP instrument is the minimization of potential contamination through the use of a physically contained sample processing unit. During extraction, the sample tube is carried to the sample processing unit. Subsequent extraction steps occur in relative isolation from other samples within the sample processing unit.
Overall, these data suggest that the CAP TNAI in combination with CTM48 has a broad measurable range and a low LOD, is highly reproducible, and agrees with HCV quantification using the M48/CTM48 test. The combined performance characteristics make the CAP TNAI/CTM48 method suitable for quantification of HCV.
Footnotes
Supported by NIAID/Social and Scientific Systems, Inc., National Institutes of Health Grant (NIH) 204VC004 and The HIV Prevention Trials Network (HPTN) sponsored by NIAID, NIDA, NIMH, and the Office of AIDS Research of the NIH, DHHS (U01-AI-068613). Reagent support was provided by Roche Diagnostics Corporation.
References
- 1.Beuselinck K, van Ranst M, van Eldere J. Automated extraction of viral-pathogen RNA and DNA for high-throughput quantitative real-time PCR. J Clin Microbiol. 2005;43:5541–5546. doi: 10.1128/JCM.43.11.5541-5546.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Forman MS, Valsamakis A. Verification of an assay for quantification of hepatitis C virus RNA by use of an analyte-specific reagent and two different extraction methods. J Clin Microbiol. 2004;42:3581–3588. doi: 10.1128/JCM.42.8.3581-3588.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jongerius JM, Sjerps M, Cuijpers HT, van Drimmelen HA, van der Poel CL, Reesink HW, Molijn MH, Peeters GA, Peeters TP, Lelie PN. Validation of the NucliSens extractor combined with the AmpliScreen HIV version 1.5 and HCV version 2.0 test for application in NAT minipool screening. Transfusion. 2002;42:792–797. doi: 10.1046/j.1537-2995.2002.00113.x. [DOI] [PubMed] [Google Scholar]
- 4.Konnick EQ, Williams SM, Ashwood ER, Hillyard DR. Evaluation of the COBAS hepatitis C virus (HCV) TaqMan analyte-specific reagent assay and comparison to the COBAS Amplicor HCV Monitor v2.0 and Versant HCV bDNA 3.0 assays. J Clin Microbiol. 2005;43:2133–2140. doi: 10.1128/JCM.43.5.2133-2140.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stelzl E, Kormann-Klement A, Haas J, Daghofer E, Santner BI, Marth E, Kessler HH. Evaluation of an automated sample preparation protocol for quantitative detection of hepatitis C virus RNA. J Clin Microbiol. 2002;40:1447–1450. doi: 10.1128/JCM.40.4.1447-1450.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fiebelkorn KR, Lee BG, Hill CE, Caliendo AM, Nolte FS. Clinical evaluation of an automated nucleic acid isolation system. Clin Chem. 2002;48:1613–1615. [PubMed] [Google Scholar]
- 7.Sizmann D, Boeck C, Boelter J, Fischer D, Miethke M, Nicolaus S, Zadak M, Babiel R. Fully automated quantification of hepatitis C virus (HCV) RNA in human plasma and human serum by the COBAS AmpliPrep/COBAS TaqMan system. J Clin Virol. 2007;38:326–333. doi: 10.1016/j.jcv.2006.12.021. [DOI] [PubMed] [Google Scholar]
- 8.Corbet S, Bukh J, Heinsen A, Fomsgaard A. Hepatitis C virus subtyping by a core-envelope 1-based reverse transcriptase PCR assay with sequencing and its use in determining subtype distribution among Danish patients. J Clin Microbiol. 2003;41:1091–1100. doi: 10.1128/JCM.41.3.1091-1100.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caliendo AM, Valsamakis A, Zhou Y, Yen-Lieberman B, Andersen J, Young S, Ferreira-Gonzalez A, Tsongalis GJ, Pyles R, Bremer JW, Lurain NS. Multilaboratory comparison of hepatitis C virus viral load assays. J Clin Microbiol. 2006;44:1726–1732. doi: 10.1128/JCM.44.5.1726-1732.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barbeau JM, Goforth J, Caliendo AM, Nolte FS. Performance characteristics of a quantitative TaqMan hepatitis C virus RNA analyte-specific reagent. J Clin Microbiol. 2004;42:3739–3746. doi: 10.1128/JCM.42.8.3739-3746.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mellor J, Hawkins A, Simmonds P. Genotype dependence of hepatitis C virus load measurement in commercially available quantitative assays. J Clin Microbiol. 1999;37:2525–2532. doi: 10.1128/jcm.37.8.2525-2532.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gelderblom HC, Menting S, Beld MG. Clinical performance of the new Roche COBAS TaqMan HCV Test and High Pure System for extraction, detection and quantitation of HCV RNA in plasma and serum. Antivir Ther. 2006;11:95–103. [PubMed] [Google Scholar]
- 13.Sarrazin C, Gartner BC, Sizmann D, Babiel R, Mihm U, Hofmann WP, von Wagner M, Zeuzem S. Comparison of conventional PCR with real-time PCR and branched DNA-based assays for hepatitis C virus RNA quantification and clinical significance for genotypes 1 to 5. J Clin Microbiol. 2006;44:729–737. doi: 10.1128/JCM.44.3.729-737.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Colson P, Motte A, Tamalet C. Broad differences between the COBAS AmpliPrep total nucleic acid isolation-COBAS TaqMan 48 hepatitis C virus (HCV) and COBAS HCV Monitor v2.0 assays for quantification of serum HCV RNA of non-1 genotypes. J Clin Microbiol. 2006;44:1602–1607. doi: 10.1128/JCM.44.4.1602-1603.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]


