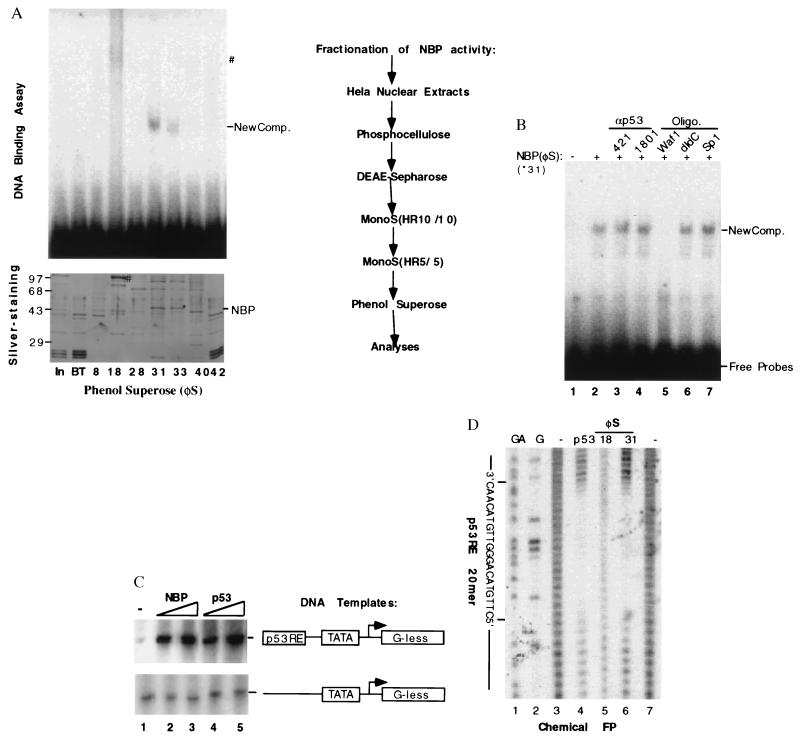
Figure 2.
Purification and functional characterization of NBP. (A) Purification steps of the NBP protein from HeLa cell nuclear extracts and analyses of the fractions from the last phenol Superose (φS) by EMSA and silver-staining SDS-PAGE. The columns used for purification of the p53RE-mediated transcription activity were listed on right. On left are shown the results from EMSA (Upper) and silver-stained SDS/10% polyacrylamide gel analyses of the peak fractions from the last column. For EMSA, 2 μl of each fraction was used & for silver-staining gel, 10 μl of each fraction was directly loaded. NBP and ∗ indicate the polypeptide band on the denatured gel, which is corresponding to the p53RE binding complexes on the EMSA gel. #, The polypeptide (Lower) that may be responsible for the smeared DNA-protein complex on the native gel (Upper). (B) The effect of anti-p53 antibodies and DNA oligomers on the p53RE-binding activity of the purified NBP. Fraction 31 (60 ng) was used in this EMSA. 0.5 μg of 421 and 1801, 0.1 μg of each oligomer as indicated on top were incubated with the purified protein 15 min before addition of the 32P-labeled p53RE DNA probe mixture into the mixture. Similar assays were conducted with anti-p73 antibodies and the mutant p53RE DNA oligomers derived from the p21 promoter. Anti-p73 antibodies and the mutant oligomers did not affect the NBP-p53RE complex (data not shown). New Comp., The NBP-p53RE complex. (C) The transcription activity of NBP in vitro. The in vitro transcription assay was conducted by using the same conditions as for Fig. 1A. Fraction 31 (60 and 120 ng) of the phenol Superose (φS) column and purified p53 (40 and 80 ng) were used in the reaction, respectively. (D) The chemical footprinting analysis of the purified NBP protein. For each lane, the DNA probes were pooled from quadruple DNA-binding reactions.