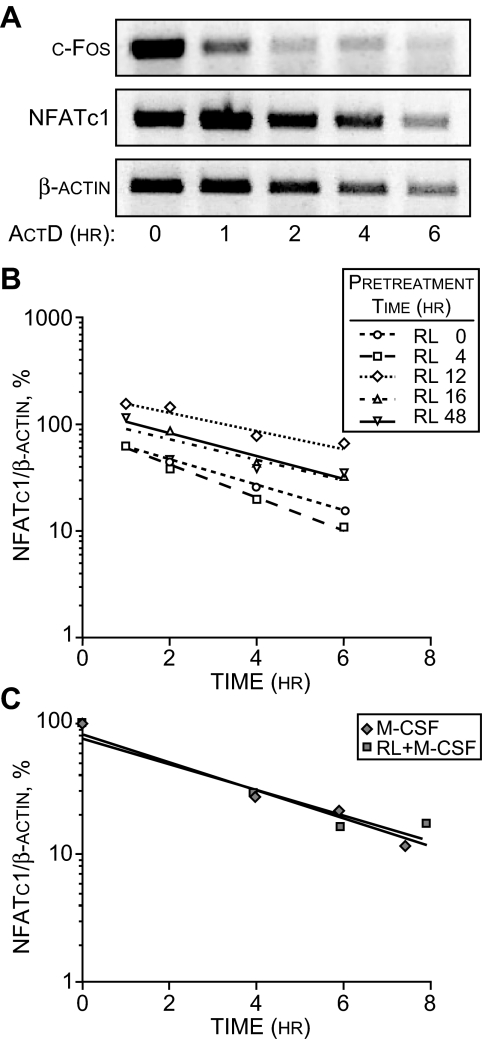
Figure 2.
The Presence of RANKL Does Not Alter NFATc1 mRNA Stability
A, NFATc1 transcripts are more stable than c-Fos mRNA. RAW264.7 were treated with sRANKL (sRL) (300 ng/ml) for 12 h to up-regulate NFATc1 mRNA and then exposed to ActD (3 μg/ml) for the periods indicated. RNA was then collected and subjected to RT-PCR analysis for β-actin (20 cycles), NFATc1 (21 cycles), or c-Fos (25 cycles) using the primers indicated in Materials and Methods. B, The stability of NFATc1 mRNA is independent of RANKL induction time in RAW264.7 cells. Cells were treated with sRANKL for increasing periods of time and exposed to ActD (3 μg/ml), and isolated RNA was subjected to qRT-PCR analysis. The amount of NFATc1 mRNA present at each time point after ActD treatment was contrasted to that before the administration of ActD, and all concentrations were normalized to the amount of β-actin mRNA present. Each determination is the average of three independent measurements. C, RANKL does not alter the stability of NFATc1 mRNA derived from primary splenic monocytes. Monocytes were isolated from C57Bl/6 mice and cultured for 2.5 d in the presence of M-CSF (10 ng/ml) alone or M-CSF (10 ng/ml) plus sRANKL (300 ng/ml). Cells were then treated with ActD (3 μg/ml), and total RNA was harvested at the times indicated in the figure. The amount of NFATc1 mRNA present at each time was determined using qRT-PCR analysis and normalized to that present before the addition of ActD. Each determination represents the average of three independent measurements.