Figure 4.
NFATc1 Autoregulates Its Expression in Developing Osteoclasts
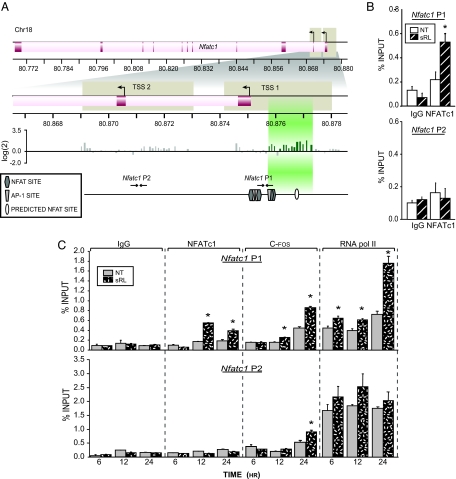
A, ChIP-chip analysis of the mouse Nfatc1 gene. RAW264.7 cells were cultured in the presence or absence of sRANKL (sRL) for 48 h and then subjected to a ChIP analysis using antibodies to NFATc1. The DNAs from sRL and control treated cells were separately amplified and labeled as outlined in Materials and Methods. The regions probed in the DNA array on chromosome 18 (Chr18) are delineated by tan boxes. The TSS and translational start sites (ATG) of Nfatc1 are shown. Signal intensity as log(cy3/cy5) ratios are shown for each probe. Peaks of NFAT binding are highlighted in green. Confirmed NFATc1 (hexagon) and AP-1 (trapezoid) binding sites as well as a predicted NFAT site (oval) are indicated for the P1 and P2 regions. The locations of the primers designed to amplify these regions are depicted with arrows and numbered according to their location relative to the P1 TSS. B, Confirmation of ChIP-chip results at promoters of Nfatc1. RAW264.7 cells were cultured in the presence or absence of sRL for 48 h and then subjected to ChIP analysis using antibodies to NFATc1 and an IgG control. The resulting coprecipitated DNA was evaluated by qRT-PCR analysis using primers capable of detecting the Nfatc1 P1 or P2. Signal intensities were normalized to their corresponding input signals. C, NFATc1 occupation of the Nfatc1 P1 region is delayed relative to the appearance of RNA pol II. RAW264.7 cells were treated with sRL for either 6, 12, or 24 h and then subjected to ChIP analysis using antibodies to NFATc1, c-Fos, RNA pol II, or IgG. The precipitated DNA was then evaluated for each of the time points as in panel B above. The data represent the average of triplicate determinations ± sem. *, P ≤ 0.05 compared with time-matched NT (no treatment) control (one-way ANOVA). These data are representative of more than 10 independent experiments.