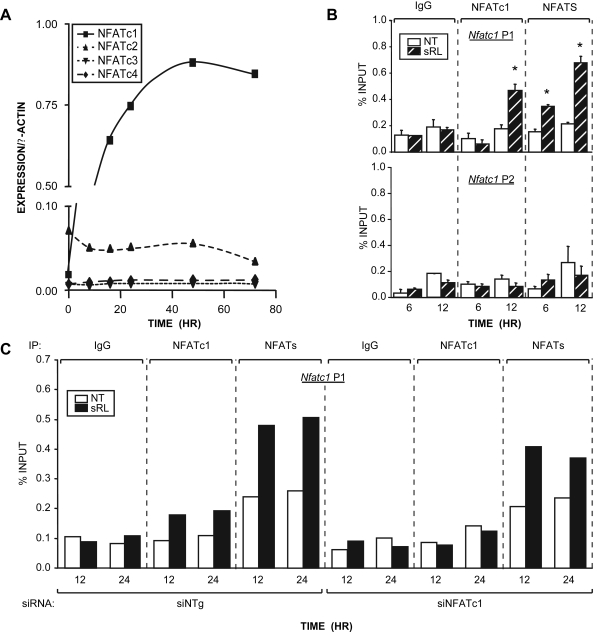
Figure 5.
NFATc2 as well as Other NFAT Proteins Participate in the Initial Upregulation of Nfatc1 expression
A, NFAT expression in RAW264.7 cells is limited to NFATc1 and NFATc2. Cells were treated with sRANKL (sRL) (300 ng/ml) for the times indicated. Total RNA was collected and subjected to RT-PCR analysis using primers designed to amplify NFATc1 (21 cycles), NFATc2 (25 cycles), NFATc3 (35 cycles), NFATc4 (35 cycles), and β-actin (21 cycles). Densitometry was performed, and the relative expression levels of the test mRNAs were normalized to β-actin levels. B, Additional NFATc members activate Nfatc1 at early times. RAW264.7 cells were treated with sRL for either 6 or 12 h and then subjected to ChIP analysis using antibodies to NFATc1, IgG, or a pan antibody to NFATc1–4. The resulting coprecipitated DNA was evaluated by qRT-PCR analysis using primers capable of detecting the Nfatc1 P1 or P2. Signal intensities were normalized to their corresponding input signals. C, Binding of other NFATc members to the Nfatc1 promoter is supported by NFATc1 knockdown. RAW264.7 cells were transfected with siRNA to knockdown expression of NFATc1 (siNFATc1) or a genome-selected nontargeted negative control siRNA (siNTg). These cells were treated with sRL for either 12 or 24 h and then subjected to ChIP analysis and evaluated as described in panel B above. The data represent the average of triplicate determinations ± sem. *, P ≤ 0.05 compared with time-matched NT control (one-way ANOVA). These data are representative of more than three independent experiments. IP, Immunoprecipitation; NT, no treatment.