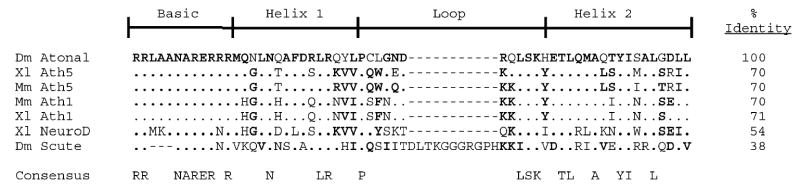
Fig. 1.
Alignment of bHLH domains for proteins analyzed in this cross-species functional comparison. ClustalW alignment of 56 (Atonal, Xath5, Math5, Math1, Xath1 NeuroD) or 64 (Scute) amino acids that comprise the basic, helix 1, loop, and helix 2 conserved domain. Consensus amino acids are indicated along the bottom, with the amino acid letter denoting perfect conservation. Dashes represent spaces introduced for optimal alignment of the basic and loop subdomains between scute and the other proteins. A “.” represents amino acids identical to Atonal, and bold type indicates those residues that are identical or similar in three or more proteins.