Abstract
Autophagy has been proposed to promote cell death during lumen formation in three-dimensional mammary epithelial acini because numerous autophagic vacuoles are observed in the dying central cells during morphogenesis. Because these central cells die due to extracellular matrix (ECM) deprivation (anoikis), we have directly interrogated how matrix detachment regulates autophagy. Detachment induces autophagy in both nontumorigenic epithelial lines and in primary epithelial cells. RNA interference-mediated depletion of autophagy regulators (ATGs) inhibits detachment-induced autophagy, enhances apoptosis, and reduces clonogenic recovery after anoikis. Remarkably, matrix-detached cells still exhibit autophagy when apoptosis is blocked by Bcl-2 overexpression, and ATG depletion reduces the clonogenic survival of Bcl-2–expressing cells after detachment. Finally, stable reduction of ATG5 or ATG7 in MCF-10A acini enhances luminal apoptosis during morphogenesis and fails to elicit long-term luminal filling, even when combined with apoptotic inhibition mediated by Bcl-2 overexpression. Thus, autophagy promotes epithelial cell survival during anoikis, including detached cells harboring antiapoptotic lesions.
INTRODUCTION
Macroautophagy (hereafter called autophagy) is an evolutionarily conserved lysosomal process where a cell degrades its own cytoplasmic contents (Levine and Klionsky, 2004). Accumulating evidence indicates that the exact role of autophagy in cell survival versus death is both stimulus and context dependent (Debnath et al., 2005; Levine and Yuan, 2005). Indeed, autophagy is well recognized as a survival mechanism during nutrient limitation; through the bulk degradation of cytoplasmic material, autophagy generates both nutrients and energy in starving cells (Levine and Klionsky, 2004). Accordingly, during nutrient starvation, inhibition of autophagy promotes apoptosis (Boya et al., 2005). In contrast, excessive autophagy has been proposed to mediate autophagic or type 2 programmed cell death (PCD). For example, L929 fibrosarcoma cells die in a caspase-independent manner involving autophagy; ATG genes are required for this death process (Yu et al., 2004). In this model, caspase inhibition induces the selective autophagic degradation of catalase, a major reactive oxygen species (ROS) scavenger, and the resulting ROS accumulation promotes type 2 PCD (Yu et al., 2006). Type 2 PCD has also been described during development, most notably, during the destruction of larval salivary glands in Drosophila (Lee and Baehrecke, 2001; Martin and Baehrecke, 2004). Presumably, the level of self-degradation becomes incompatible with life, leading to cellular demise.
Both autophagy and apoptosis are observed during lumen formation in three-dimensional (3D) epithelial cultures in vitro. When cultured on reconstituted basement membrane, MCF-10A cells, a nontransformed human mammary epithelial cell line, form spherical structures (termed “acini”) in which a layer of polarized epithelial cells surrounds a hollow lumen, resembling glandular epithelium in vivo (Debnath et al., 2003). The detailed analysis of 3D morphogenesis reveals the presence of two populations in developing acini: an “outer” cell layer directly attached to the extracellular matrix (ECM), and a centrally located subset of “inner” cells lacking ECM contact. Lumen formation involves the selective apoptosis of these central cells; caspase-3 cleavage is observed in dying cells as the lumen hollows (Debnath et al., 2002). However, inhibiting apoptosis by the ectopic expression of either Bcl-2 or Bcl-xL only delays lumen clearance for a few days. Ultimately, these acini do form hollow lumen. Electron microscopic analysis reveals that numerous autophagic vacuoles are present in the central cells of developing acini (Debnath et al., 2002; Underwood et al., 2006). Notably, the central cells in Bcl-2–expressing structures also exhibit extensive autophagy (Debnath et al., 2002).
Based on these initial results, autophagy has been proposed to promote luminal clearance by autophagic death (Debnath et al., 2002; Mills et al., 2004). Notably, follow-up studies indicate that a tumor necrosis factor family ligand, Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) induces autophagy in epithelial cells and that TRAIL inhibition promotes luminal filling when combined with Bcl-xL–mediated inhibition of apoptosis (Debnath et al., 2002; Mills et al., 2004). Although these results suggest that both apoptosis and autophagy are required for luminal cell death and clearance, these studies are correlative (Debnath et al., 2005). Importantly, epithelial cells critically depend on integrin-mediated cell adhesion to ECM for proper growth and survival; upon detachment, epithelial cells undergo apoptotic cell death, termed anoikis (Frisch and Francis, 1994). Thus, one can alternatively hypothesize that autophagy is induced during anoikis as a survival strategy to mitigate the stresses of ECM detachment. Because protection from anoikis is thought to promote tumor cell survival in vivo and to contribute to luminal filling in glandular structures, we interrogated whether autophagy is induced in epithelial cells due to ECM detachment. We also tested whether autophagy regulators (ATGs) promote epithelial cell survival, versus type 2 autophagic cell death, during anoikis and 3D lumen formation.
MATERIALS AND METHODS
Cell Culture
MCF-10A cells were cultured as described previously (Debnath et al., 2003). Primary human mammary epithelial cells (1°HMECs) were obtained from Cambrex (East Rutherford, NJ) and cultured in Mammary Epithelial Cell Medium (MEGM) (Cambrex). For epidermal growth factor (EGF) withdrawal studies, EGF was omitted as an exogenous supplement from either MCF-10A or MEGM growth media. Madin-Darby canine kidney (MDCK)-2 cells (gift from Dr. K. Matlin, University of Cincinnati, Cincinnati, OH) and both wild-type and ATG5−/− simian virus 40-immortalized mouse embryonic fibroblasts (gift from Dr. N. Mizushima, Tokyo Medical and Dental University, Tokyo, Japan) were grown in DMEM (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum, penicillin, and streptomycin.
Antibodies and Chemicals
A peptide corresponding to the N terminus common to human, mouse, and rat MAP1LC3 was used to create α-LC3 rabbit polyclonal antibody; the whole sera was used at 1:500–1:1000 for immunoblotting. In mammary cells, we noticed that LC3-I is not efficiently detected with this antibody when low amounts of protein lysates are analyzed. (e.g., see Figure 3B). Other antibodies used included the following: α-ATG12 (Zymed Laboratories, South San Francisco, CA); α-ATG5 (gift from N. Mizushima, Tokyo Medical and Dental University); α-ATG7 (Santa Cruz Biotechnology, Inc., Santa Cruz, CA); α-Beclin (ATG6) (BD Biosciences, San Jose, CA); α-Bim (Prosci, Poway, CA); α-cleaved caspase-3 (Cell Signaling Technology, Danvers, MA); α-catalase (Calbiochem, San Diego, CA); α-enhanced epidermal growth factor receptor (EGFR) (Cell Signaling Technology); α-laminin 5 (Chemicon International, Temecula, CA); α-α-tubulin (Sigma-Aldrich, St. Louis, MO); α-phospho-AMP-activated protein kinase (AMPK), α-AMPKα, α-P-eukaryotic initiation factor 2α (eIF2α), and eIF2α (Cell Signaling Technology); α-phopho-ERK1/2 (BioSource International. Camarillo, CA); and α-extracellular signal-regulated kinase (ERK)1/2 (Zymed Laboratories). For function-blocking studies, α-β1 integrin subunit (A2B2) was from C. Damsky (University of California San Francisco, San Francisco, CA) and α-E-cadherin (DECMA) and the rat immunoglobulin (Ig)G isotype control were from Sigma-Aldrich. Chemicals utilized included poly(2-hydroxyethyl methacrylate) (poly-HEMA), 3-methyadenine, bafilomycin A, chloroquine, E64d, and pepstatin A (all from Sigma-Aldrich).
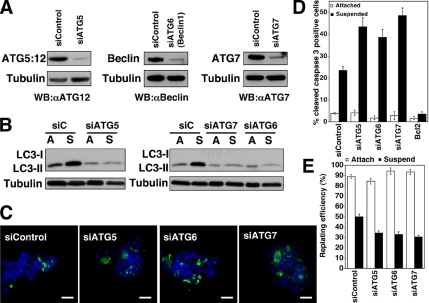
Figure 3.
ATG depletion enhances apoptosis in ECM-detached cells. (A) MCF-10A cells transfected with pooled nontargeting control (siControl) or siRNA oligonucleotides against ATG5, ATG6 (Beclin 1), or ATG7 were lysed and immunoblotted with α-ATG12 to detect the ATG5:12 complex, α-Beclin, α-ATG7, and α-tubulin. (B) Cells transfected with the indicated siRNAs were grown attached (A) or suspended (S) for 24 h, lysed, and subjected to α-LC3 and α-tubulin immunoblotting. LC3-I detection was minimal in this experiment. (C) Confocal images of MCF-10A cells transfected with the indicated siRNAs, suspended for 24 h, fixed, and immunostained with α-cleaved caspase-3 antibody (green). Nuclei were stained with 4,6-diamidino-2-phenylindole (DAPI) (blue). Bars, 20 μm. (D) Percentage of cleaved caspase-3–positive cells in MCF-10A cells transfected with the indicated siRNAs and grown attached (white) or suspended (black) for 24 h. Results are the mean ± SEM of five or more independent experiments; in each experiment, 200–300 cells were analyzed. (E) Clonogenic replating efficiency of cells transfected with the indicated siRNAs after 48-h attachment (white) or suspension (black). Replating efficiency was calculated as the number of colonies formed divided by the number of cells originally replated. Results are the mean ± SEM from three to five independent experiments.
Generation of Stable Lines
The following retroviral vectors for stable gene expression have been described previously: pBABEpuro-Bcl-2, pBABEneo-Bcl-2, pBABEhygro-Bcl-xL, and pLPCX-EGFR (Debnath et al., 2002; Reginato et al., 2003). Rat green fluorescent protein fused to the mammalian ATG8 orthologue, microtubule-associated protein light chain 3 (GFP-LC3), was subcloned from pEGFPC1-LC3, a gift from T Yoshimori (National Institute of Genetics, Mishima, Japan), into the SnaB1 and Sal1 sites of pBABEpuro (Kabeya et al., 2000). Vesicular stomatitis virus G protein-pseudotyped retroviruses were generated, and MCF-10A lines were infected and selected as described previously (Debnath et al., 2003).
RNA Interference
Pooled small interfering RNA (siRNA) oligonucleotides (SMARTpool) against ATG5, ATG6 (Beclin 1), or ATG7 were purchased from Dharmacon RNA Technologies (Lafayette, CO). For siRNA transfection, cells were seeded at 100,000/well in six-well tissue dishes, and they were transfected with 50–100 nM of the pooled oligonucleotide mixture by using Oligofectamine (Invitrogen) following manufacturer's protocols. The transfection media were removed, and cells were allowed to recover in complete growth media for 36–48 h before use in experiments. The sense sequences of the individual duplexes directed against ATG5, 6, and 7 are as follows: ATG5 (NM_004849): GGAAUAUCCUGCAGAAGAAUU, CAUCUGAGCUACCCGGAUAUU, GACAAGAAGACAUUAGUGAUU, and CAAUUGGUUUGCUAUUUGAUU; BECN1(ATG6) (NM_003766): CUAAGGAGCUGCCGUUAUAUU, GGAUGACAGUGAACAGUUAUU, UAAGAUGGGUCUGAAAUUUUU, and GCCAACAGCUUCACUCUGAUU; and ATG7 (NM_006395): CCAAAGUUCUUGAUCAAUAUU, GAUCAAAGGUUUUCACUAAUU, GAAGAUAACAAUUGGUGUAUU, and CAACAUCCCUGGUUACAAGUU.
DNA oligonucleotides encoding short hairpin RNA (shRNA) sequences against ATG5 were chemically synthesized (Dharmacon RNA Technologies), annealed, and subcloned into BglII and XhoI sites of the pSRpuro retroviral vector to express shRNAs under the control of the H1 promoter. The target sequence of two small hairpin RNAs against ATG5 are as follows: shATG5hp1, GGATGAGATAACTGAAAGG; and shATG5hp2, GGCATTATCCAATTGGTTT.
Lentiviral particles expressing small hairpin RNAs against ATG7 were purchased from Sigma-Aldrich (Mission shRNA). The target sequence of the two hairpins used were shATG7hp1 (NM_006395; TRCN0000007584), GCCTGCTGAGGAGCTCTCCAT; and shATG7hp2 (TRCN0000007587), CCCAGCTATTGGAACACTGTA.
Substratum Detachment Assays
Tissue culture plates were coated with 6 mg/ml poly-HEMA in 95% ethanol, and then they were incubated at 37°C for several days until dry. Cells were plated on poly-HEMA–coated plates at a density of 100,000–250,000 cells/well in their appropriate complete growth medium. To analyze LC3-II turnover, the lysosomal inhibitors E64d and pepstatin A were added directly to the culture media at 10 μg/ml at 2–4 h before harvest. To quantify apoptosis, cells were suspended or attached for 24 h, collected, fixed, and stained with cleaved caspase-3 antibody by using protocols described previously (Debnath et al., 2003). To enumerate the percentage of cells positive for cleaved caspase-3, 200–300 cells were counted for each condition, and each experiment was repeated at least five times. For electron microscopy of detached cells, samples were processed at the Harvard Medical School Electron Microscopy core facility by using protocols described previously, and they were imaged using a JEOL-60kV microscope (Debnath et al., 2002; Mills et al., 2004).
Analysis of Punctate GFP-LC3
MCF-10A or 1°HMECs stably expressing GFP-LC3 were grown overnight attached on 2% Matrigel-coated coverslips before EGF withdrawal or starvation with Hank's balanced salt solution (HBSS) for the indicated times, or grown suspended for the indicated times on poly-HEMA–coated plates. Cells were fixed with 2% paraformaldehyde, washed several times with phosphate-buffered saline (PBS), mounted using Immunomount (Thermo Electron, Waltham, MA), and analyzed at 20°C by widefield immunofluorescent microscopy by using the 63× (1.4 numerical aperture [NA]) or 100× (1.3 NA) objectives of an Axiovert 200 microscope (Carl Zeiss, Thornwood, NY) equipped with a Spot RT camera (Diagnostic Instruments, Sterling Heights, MI) and mercury lamp; images were acquired using MetaMorph (version 6.0) software (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom).
Clonogenic Replating Assays
Cells (100,000) were grown attached or suspended on poly-HEMA–coated dishes for 48 h, collected, and treated with 0.25% trypsin-EDTA at 37°C for 15 min to generate single cell suspensions. Cells were counted and replated in complete growth media at 100 cells/well onto 12-well tissue culture plates. Colonies were grown out for 5 d, fixed with 2% paraformaldehyde, and stained with 0.2% crystal violet in PBS. The number of colonies was enumerated, and replating efficiency was calculated as the number of colonies growing out divided by the original number of cells plated. For each experiment, four to six replicates were performed for each condition tested, and each experiment was repeated three to five times.
3D Morphogenesis Assays
3D assays were carried out and processed for ethidium bromide (EtBr) staining or confocal microscopy as described previously (Debnath et al., 2003). Where indicated, 10 mM 3-methyladenine, 20 nM bafilomycin A, or 20 μM chloroquine was added on day 6.
Immunoblot Analysis
Attached or suspended cells were lysed in radioimmunoprecipitation assay (1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS, 25 mM Tris, pH 7.6, 150 mM NaCl, 10 mM NaF, 10 mM β-glycerophosphate, 1 mM Na3VO3, and 10 nM calyculin A) plus protease inhibitors. Lysates were cleared by centrifugation for 15 min at 4°C, boiled in SDS sample buffer, resolved usingSDS-polyacrylamide gel electrophoresis (PAGE), and transferred to polyvinylidene difluoride membrane. The membranes were blocked in PBS + 0.1% Tween 20 with 5% nonfat dry milk, incubated with the primary antibodies indicated overnight at 4°C, washed, incubated with horseradish peroxidase-conjugated secondary antibodies, and analyzed by enhanced chemiluminescence.
Immunofluorescence and Confocal Microscopy
Widefield immunofluorescence imaging was performed on an Axiovert 200 microscope (Carl Zeiss) equipped with a mercury lamp. Images were acquired at 20°C with a 20× (0.4 NA) objective with a Spot RT charge-coupled device (CCD) camera (Diagnostic Instruments) and MetaMorph (version 6.0) software. Confocal imaging was performed at 20°C, by using an Axiovert 200 (Carl Zeiss) equipped with a Yokogawa CSU-10 spinning disk, an argon laser (488 line), and two solid-state diode lasers (405 and 546 lines). Images were acquired using a 40× (1.3 NA) objective with an Andor iXON CCD camera and MetaMorph (version7.0) software. Images were color-combined in MetaMorph (version 6.0).
RESULTS
Substratum Detachment Induces Autophagy in Epithelial Cells and Fibroblasts
To monitor autophagosome formation, we created MCF-10A cells and 1°HMECs that stably express GFP-LC3. During autophagy, LC3 is modified with the lipid phospatidylethanolamine (PE) through an ubiquitin-like conjugation process, upon which it specifically relocates to early autophagosomes; accordingly, the relocation of GFP-LC3 to easily visualized “puncta” has emerged as a powerful technique to monitor autophagosome formation (Kabeya et al., 2000). Because transient GFP-LC3 transfection can produce overexpression artifacts, we developed retroviral vectors (pBABE) encoding GFP-LC3, and we generated stable pools ectopically expressing these fusions (Kuma et al., 2007). In these cells, we confirmed that GFP-LC3 relocates to autophagosomes during HBSS-induced nutrient starvation (Figure 1A).
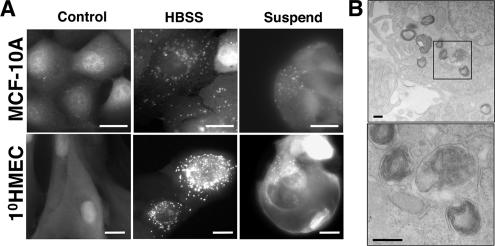
Figure 1.
Substratum detachment induces autophagosome formation in human mammary epithelial cells. (A) MCF-10A cells or 1°HMECs expressing GFP-LC3 were grown attached in full growth media for 48 h (control, left column), starved with Hank's balanced salt solution for 3 h (HBSS, center), or grown detached on poly-HEMA–coated plates in complete growth media for 48 h (suspend, right). Bars, 20 μm. (B) MCF-10A cells grown detached for 24 h were analyzed by transmission electron microscopy. The boxed area in upper panel is enlarged in lower panel. Bars, 200 nm.
Integrin-mediated cell adhesion to ECM is critical for proper growth and survival (Meredith et al., 1993; Frisch and Francis, 1994). Thus, we hypothesized that the lack of ECM contact would induce autophagy in mammary epithelial cells during anoikis, a type of apoptotic cell death that epithelial cells undergo when detached from an underlying substratum for extended periods (Frisch and Francis, 1994). To induce anoikis, we incubated cells on poly-HEMA–coated tissue culture dishes to prevent cell attachment. Although epithelial cells form clusters in these conditions due to increased cell–cell contact, they ultimately undergo apoptosis due to the lack of ECM contact (Frisch, 1999; Reginato et al., 2003). We found that substratum detachment strongly induced GFP-LC3 puncta in both MCF-10A cells and 1°HMECs (Figure 1A). In addition, using electron microscopy, we corroborated the presence of autophagic vacuoles; double membrane vacuoles containing cytoplasmic contents were observed in detached cells (Figure 1B).
In contrast to cells grown as monolayers (Supplemental Figure S1), we were unable to reliably quantify the number of GFP-LC3 puncta per cell by using image analysis software, due to the clustering of cells during anoikis. As a result, to measure autophagy during anoikis, we examined the PE-lipid conjugation of endogenous LC3 in cell lysates from detached cells; the PE-modification of LC3 results in a faster migrating isoform (called LC3-II) that can be detected by immunoblotting (Kabeya et al., 2000). Increased PE-modified LC3-II was observed in MCF-10A cells after 24 h of detachment (Figure 2A). Because LC3-II is subject to lysosomal degradation during autophagy, increased LC3-II levels do not always accurately reflect full-fledged autophagic degradation (Tanida et al., 2005). To more precisely assess autophagic flux, we assessed LC3-II levels in both attached controls and detached cells treated with lysosomal cathepsin inhibitors E64d and pepstatin A. These experiments confirmed increased LC3-II turnover during ECM detachment (Figure 2B, E64d/pepA + lanes). Interestingly, in attached 1°HMECs, we found high baseline levels of LC3-II that were reduced upon detachment (Figure 2C, E64d/pepA − lanes). Nonetheless, we once again observed increased LC3-II in the presence of these inhibitors (Figure 2C, E64d/pepA + lanes). Altogether, these experiments confirmed increased LC3-II turnover, consistent with increased autophagic degradation in MCF-10A cells and 1°HMECs during substratum detachment.
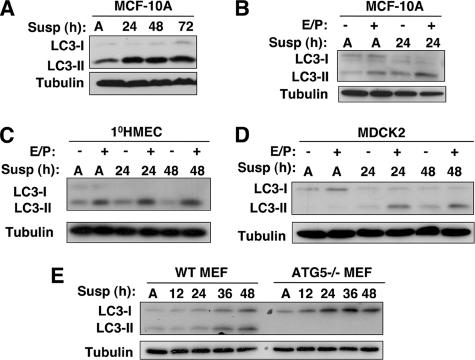
Figure 2.
Detachment-induced autophagy in epithelial cells and fibroblasts. (A) Lysates from MCF-10A cells grown attached (A) for 48 h or suspended for the indicated times were immunoblotted with α-LC3 and α-tubulin. (B and C) MCF-10A and 1°HMEC lysates grown attached (A) or suspended for the indicated times were subject to α-LC3 and α-tubulin immunoblotting. When indicated by E/P+, E64d and pepstatin A (10 μg/ml each) were added directly to the culture 4 h before lysis. (D) Cell lysates collected from MDCK2 cells grown attached for 48 h (A) or suspended for the indicated times in complete growth media. When indicated by E/P+, E64d and pepstatin A (10 μg/ml each) were added directly to the culture 4 h before lysis. (E) Wild-type or ATG5−/− MEFs were grown attached for 48 h or detached in complete media for the indicated times. All lysates were subject to immunoblotting with α-LC3 and α-tubulin antibodies.
Furthermore, we examined whether autophagy was induced in other cells during matrix detachment. We observed increased LC3-II turnover during ECM detachment in MDCK2 cells (Figure 2D), a nontumorigenic epithelial cell line susceptible to anoikis (Rytomaa et al., 2000). Finally, we found increased LC3-II formation during the detachment of mouse embryonic fibroblasts (MEFs), which do not undergo apoptosis during matrix detachment, but critically depend on integrin-based signals for proper cell growth and proliferation (Miranti and Brugge, 2002). LC3-II conversion was potently inhibited in autophagy deficient ATG5−/− MEFs, although increased unmodified LC3 (LC3-I) levels were observed (Figure 2E) (Kuma et al., 2004). Thus, autophagy is a general response to substratum detachment in various epithelial cells and in mouse fibroblasts.
We next sought to clarify whether the autophagy induced by substratum detachment could more directly result from reduced ECM or integrin-mediated signals. When exogenous laminin-rich reconstituted basement membrane (2%) was added to suspended cells, LC3-II formation decreased (Supplemental Figure S2A). We speculate that these rescue experiments did not completely inhibit autophagy because exogenous basement membrane addition fails to restore the mechanical forces critical for proper integrin function (Katsumi et al., 2004). To further interrogate the role of integrin receptor engagement, we examined autophagy in cells incubated with function-blocking antibodies directed against the β1 integrin subunit, a critical regulator of anoikis in epithelial cells (Reginato et al., 2003). Antibody-mediated blockade of β1 integrin (using A2B2) increased LC3-II formation (Supplemental Figure S2B). In contrast, we did not observe the induction of LC3-II upon inhibiting E-cadherin–mediated cell–cell adhesion by using a blocking antibody (DECMA) (Supplemental Figure S2B). Thus, a laminin-rich matrix or reduced β1 integrin function can modulate autophagosome formation in detached epithelial cells.
ATG Depletion Enhances Apoptosis in Detached Cells
Autophagy is tightly regulated by a limited number of highly conserved molecules called ATGs (Klionsky et al., 2003). Because ATG depletion inhibits autophagy, we used siRNA oligonucleotide pools targeting human ATG5, 6, and 7 to reduce autophagy during matrix detachment. We confirmed siRNA-mediated reduction of endogenous ATG5, ATG6 (Beclin1), and ATG7 in MCF-10A cells by immunoblotting (Figure 3A), which all produced significant decreases in LC3-II during anoikis (Figure 3B). In parallel, we quantified how ATG reduction affects GFP-LC3 puncta formation during HBSS starvation; cells with reduced ATG5 exhibited an 80–90% reduction in GFP-LC3 dots per cell, whereas cells with reduced ATG6 and 7 exhibited milder inhibition (Supplemental Figure S1A). Overall, these results indicated that silencing of multiple ATGs results in reduced autophagy. Cells with reduced ATGs were subject to matrix detachment for 24 h, and the levels of apoptosis were measured by enumerating the percent of cells within a culture with positive staining for cleaved caspase-3 (Figure 3C). In cells with reduced ATGs, we observed a 1.5- to 2.0-fold increase in cleaved caspase-3 in suspended cells; in contrast, no differences in apoptosis were observed in attached controls (Figure 3D). Notably, these proapoptotic effects were observed with the silencing of multiple independent ATGs, arguing against nonspecific effects due to any individual ATG knockdown. Induction of the proapoptotic BH3-family protein Bim critically mediates apoptosis in detached mammary epithelial cells (Reginato et al., 2003). As a result, we examined whether ATG depletion amplified Bim induction during anoikis; however, we did not observe changes in Bim protein levels in cells with reduced ATGs versus controls (Supplemental Figure S3).
Finally, to more directly interrogate how autophagy regulates cell viability after anoikis, we assayed clonogenic potential after ECM detachment. After 48 h of matrix detachment, single cell suspensions were generated from control and ATG-depleted cells and replated to assess clonogenic recovery and colony formation. In cells subjected to ECM detachment, we found decreased colony formation in ATG-depleted cultures versus controls; in contrast, reduced clonogenicity was not observed in attached cells with reduced ATGs (Figure 3E). Thus, similar to cells undergoing nutrient starvation, the inhibition of autophagy promotes apoptosis and decreases the survival of cells deprived of ECM contact (Boya et al., 2005).
Detachment-induced Autophagy Promotes the Survival of Cells Expressing Bcl-2
Our previous studies of lumen formation in Bcl-2–overexpressing acini indicate that apoptosis inhibition does not affect autophagy induction in cells occupying the lumen (Debnath et al., 2002; Mills et al., 2004). Accumulating evidence indicates that depending on cell type, context, or both, Bcl-2 inhibits autophagy (Pattingre et al., 2005), simulates autophagy and ATG-dependent cell death (Shimizu et al., 2004), or permits autophagy to develop by inhibiting apoptosis (Degenhardt et al., 2006). To distinguish among these possibilities, we examined the induction of autophagy during matrix detachment in cells expressing antiapoptotic Bcl-2 family proteins. We observed no significant changes in GFP-LC3 puncta development between control versus Bcl-xL–expressing cells (Figure 4A). Similarly, by immunoblot analysis, we found no significant differences in PE-modified LC3-II within Bcl-2–expressing cells during anoikis (Figure 4B). These results indicate that antiapoptotic Bcl-2 proteins do not inhibit detachment-induced autophagy in mammary epithelial cells.
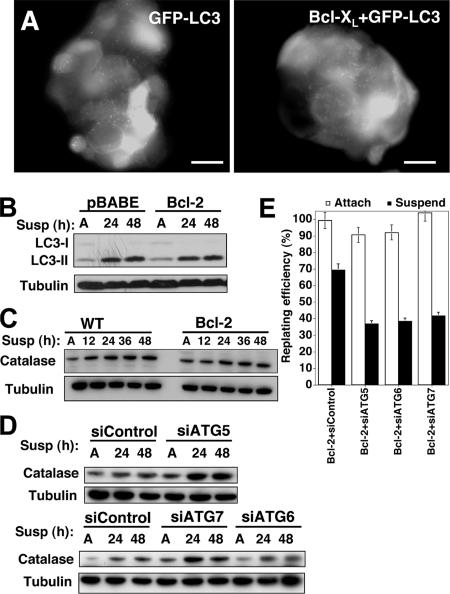
Figure 4.
Detachment-induced autophagy in Bcl-2 expressing cells. (A) Punctate GFP-LC3 in detached MCF-10A cells stably expressing GFP-LC3 or coexpressing GFP-LC3 + Bcl-xL suspended for 24 h. Bar, 20 μm. (B) Control (BABE) or Bcl-2–expressing cells were grown attached (A) for 48 h or suspended for the indicated times, lysed, and subject to immunoblotting with α-LC3 and α-tubulin antibodies. (C) Wild-type and Bcl-2–expressing cells were grown attached (A) or suspended for the indicated times, lysed, and subject to immunoblotting with α-catalase and α-tubulin antibodies. (D) Cells transfected with the indicated siRNAs were grown attached (A) or suspended for the indicated times, lysed, and subject to immunoblotting with α-catalase and α-tubulin antibodies. (E) Clonogenic replating efficiency of Bcl-2–expressing cells transfected with the indicated siRNAs after 48 h attachment (white) or suspension (black). Replating efficiency was calculated as described previously. Results are the mean ± SEM from three to five independent experiments.
When caspases are pharmacologically inhibited, catalase, a major reactive oxygen species (ROS) scavenger is selectively degraded by autophagy, which has been demonstrated to mediate type 2 autophagic death (Yu et al., 2006). Thus, to investigate the possibility that autophagy regulated type 2 cell death during ECM detachment, we examined the protein levels of catalase during anoikis. However, we observed slightly increased, rather than decreased, catalase protein levels in both wild-type and Bcl-2–expressing cells (Figure 4C). Thus, we conclude that catalase depletion does not mediate death during ECM detachment, even when apoptosis is inhibited by Bcl-2. Interestingly, we observed increased catalase protein levels in ATG knockdown cells undergoing anoikis compared with controls (Figure 4D). Although these results support that autophagy directly mediates catalase degradation during anoikis, increased catalase may arise as a secondary consequence of increased reactive oxygen species levels in detached ATG knockdown cells.
Growing evidence indicates that autophagy promotes the viability of cells unable to undergo apoptosis. When lymphocytes doubly deficient for Bax and Bak are deprived of a critical growth factor, interleukin-3, the inhibition of autophagy elicits a rapid cell death associated with reduced ATP levels (Lum et al., 2005). Similarly, in cells unable to undergo apoptosis due to Bcl-2 or Bcl-xL expression, Beclin (ATG6) depletion impairs the induction of autophagy during ischemia and reduces cell viability (Degenhardt et al., 2006). Based on these previous studies, we hypothesized that autophagy may contribute to the survival of Bcl-2–expressing cells during matrix detachment. We measured the clonogenic viability of Bcl-2–expressing cells after 48 h of detachment. Similar to our results in wild-type cells, the knockdown of ATG5, 6, or 7 resulted in decreased colony formation when compared with Bcl-2–expressing controls (Figure 4E). As before, no differences in clonogenicity were observed in attached cells. Thus, detachment-induced autophagy promotes the viability of cells overexpressing the antiapoptotic protein Bcl-2.
Autophagy Is Induced during Epidermal Growth Factor Withdrawal
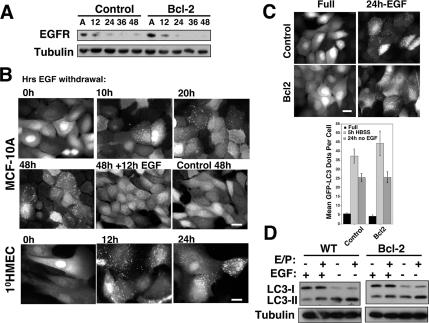
Substratum attachment and integrin-mediated cell adhesion are essential for the proper activation of growth factor receptor pathways (Miranti and Brugge, 2002). Specifically, EGFR, a growth factor receptor important for epithelial cell survival and proliferation, is significantly reduced in a variety of epithelial cells during anoikis (Reginato et al., 2003). When we examined EGFR levels in Bcl-2–expressing cells, we found that these cells exhibited EGFR down-regulation similar to wild-type controls (Figure 5A). Thus, we reasoned that even though Bcl-2–expressing cells were protected from apoptosis, they would still exhibit other biological consequences associated with EGFR down-regulation during ECM detachment. Moreover, we postulated that detachment-induced autophagy in both wild type and Bcl-2–expressing cells was at least partially due to reduced EGFR expression and pathway activation.
Figure 5.
EGF Withdrawal induces autophagy. (A) Lysates from pBABE control and Bcl-2–expressing MCF-10A cells grown attached (A) or suspended for the indicated times were subject to immunoblotting with α-EGFR and α-tubulin. (B) GFP-LC3–expressing MCF-10A cells or 1°HMECs were grown in media lacking EGF for the indicated times. Twelve hours of EGF readdition reverses LC3 puncta formation. (C) GFP-LC3 puncta in pBABE control and Bcl-2–expressing cells after 24 h of EGF withdrawal. Bar, 20 μm. Bottom, quantification of GFP-LC3 dots per cell in control and Bcl-2 cells grown in complete growth media (black), HBSS starved for 5 h (light gray), or EGF withdrawn for 24 h (dark gray). Results are the mean ± SEM enumerated from 40 to 60 individual cells using MetaMorph (GE Healthcare) software. (D) Wild-type and Bcl-2–expressing cells were grown in complete media or media lacking EGF for 24 h, lysed, and subject to immunoblotting with α-LC3 and α-tubulin.
To test this hypothesis, we first assessed whether EGF depletion was sufficient to induce autophagy in mammary cells when grown in attached conditions. Indeed, we discovered that EGF withdrawal induced autophagy in both MCF-10A cells and 1°HMECs (Figure 5B). Importantly, autophagy during EGF withdrawal ensued despite the presence of other growth factors, including insulin, hydrocortisone, and serum. Notably, we observed that autophagy was a reversible process; upon EGF readdition, GFP-LC3 puncta disappeared within 12 h (Figure 6B). After 24 h of EGF depletion, a fourfold increase in GFP-LC3 puncta was observed in MCF-10A cells. Moreover, we did not detect reduced levels of punctate GFP-LC3 in cells ectopically expressing Bcl-2, in contrast to ATG depletion during EGF withdrawal (Figure 5C and Supplemental Figure S1). Furthermore, during EGF withdrawal in both control and Bcl-2–expressing cells, we observed increased LC3-II turnover in the presence of E64d and pepstatin A (Figure 5D). Overall, these results indicated that EGF withdrawal was sufficient to induce autophagy in both wild-type and Bcl-2–expressing mammary epithelial cells.
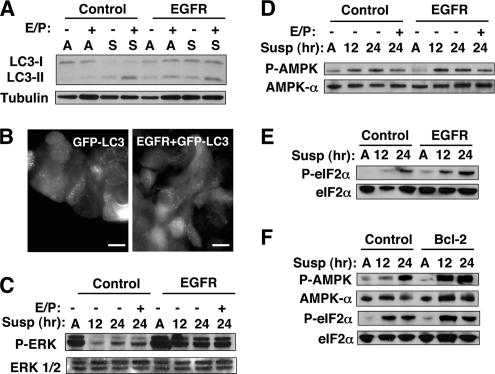
Figure 6.
Detachment-induced autophagy in cells overexpressing EGFR. (A) Control (LPCX) and EGFR-expressing cells were grown attached (A) or suspended (S) for 24 h, lysed, and subject to immunoblotting with α-LC3 and α-tubulin. (B) Punctate GFP-LC3 in detached MCF-10A cells stably expressing GFP-LC3 or GFP-LC3 + EGFR suspended for 24 h. Bar, 20 μm. (C) Control (LPCX) and EGFR-overexpressing cells grown attached (A) or suspended for the indicated times, were lysed and subject to immunoblotting with α-P-ERK1/2, α-ERK1 + 2 (D) Control (LPCX) and EGFR-overexpressing cells grown attached (A) or suspended for the indicated times, lysed, and subjected to immunoblotting with α-P-AMPK, and α-AMPKα. (E) Control (LPCX) and EGFR-overexpressing cells grown attached (A) or suspended for the indicated times, lysed, and subjected to immunoblotting with α-P-eIF2α and α-eIF2α. (F) Control (pBABE) and Bcl-2–expressing cells were grown attached of suspended for the indicated times, lysed, and subjected to immunoblotting for the indicated antibodies. In A, C, and D, the E/P + lanes indicate E64d and pepstatin A added directly to the culture 4 h before lysis.
We then tested whether the ectopic overexpression of EGFR was sufficient to inhibit detachment-induced autophagy. Surprisingly, we found that stable EGFR overexpression did not significantly inhibit LC3-II formation and turnover during matrix detachment; in addition, we observed the robust punctate GFP-LC3 in suspended EGFR-overexpressing cells (Figure 6, A and B). Based on this result, we hypothesized that although the loss of EGFR driven signals may contribute to autophagy, additional EGFR-independent pathways also positively regulated autophagy during anoikis. Thus, we probed whether specific survival and stress signaling pathways were regulated by EGFR during anoikis, whereas others were not. Indeed, we confirmed that EGFR-overexpressing cells exhibited potent and sustained activation of certain survival pathways during matrix detachment, most notably, the ERK/mitogen-activated protein kinase pathway, previously demonstrated to inhibit apoptosis (Figure 6C; Reginato et al., 2003). In contrast, we uncovered that several other stress response pathways were not inhibited by EGFR overexpression. First, during anoikis, we observed high levels of activation of the energy sensor AMPK in both control and EGFR-overexpressing cells (Figure 6D). Increasing evidence indicates that AMPK positively regulates autophagy in mammalian cells (Meley et al., 2006; Hoyer-Hansen et al., 2007; Liang et al., 2007). In addition, we found the increased phosphorylation of eukaryotic initiation factor 2α on serine 51 (P-eIF2α), a stress-regulated translational suppressor that up-regulates autophagy during nutrient starvation and endoplasmic reticulum (ER) stress (Talloczy et al., 2002; Kouroku et al., 2007). Similar to AMPK activation, the phosphorylation of eIF2α was not reduced in detached EGFR-expressing cells (Figure 6E). Remarkably, we also found increased AMPK activation and phosphorylated eIF2α in Bcl-2–expressing cells during ECM detachment (Figure 6F). Hence, even though EGFR and Bcl-2 protect detached cells from apoptosis, multiple stress pathways continue to be potently activated in cells overexpressing these prosurvival molecules.
Increased Luminal Cell Death in 3D Acini on Chemical Autophagy Inhibition
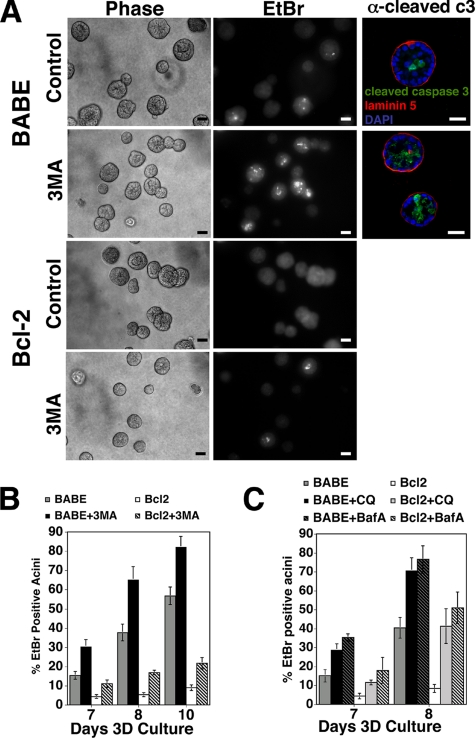
Because anoikis is a principal contributor to the selective death of central cells during lumen formation, we hypothesized that the autophagic vacuoles observed in the lumen during morphogenesis were mitigating the stresses of ECM detachment. To initially test this hypothesis, we measured the rates of luminal cell death in acini following acute pharmacological inhibition of autophagy. We treated acini with 3-methyladenine (3-MA), an established pharmacological inhibitor of early autophagosome formation. Acini derived from wild-type and Bcl-2–expressing MCF-10A cells were 3D cultured for 6 d; at this time point, minimal luminal cell death is observed (Debnath et al., 2002). On day 6, cultures were treated with 3-MA, and cell death was measured on subsequent days by staining with EtBr, a DNA-intercalating dye only incorporated into dying cells. We observed increased EtBr staining within the centers of acini treated with 3-MA (Figure 7A), which correlated with increased cleaved-caspase-3–positive cells in the lumens of individual wild-type structures (Figure 7A). When we enumerated the percentage of EtBr-positive acini in both control and 3-MA–treated cultures, we found increased luminal cell death in cultures treated with this autophagy inhibitor (Figure 7B). Similar results were obtained when day 6 acini were acutely treated with the lysosomal inhibitors chloroquine (CQ) or bafilomycin A (BafA) (Figure 7C). Finally, although Bcl-2 potently suppresses luminal cell death, increased numbers of EtBr-positive acini were observed in Bcl-2–expressing cultures treated with both autophagy and lysosomal inhibitors (Figure 7, A–C).
Figure 7.
Increased luminal cell death in 3D acini upon chemical autophagy inhibition. (A) Indicated cell types were cultured for 6 d on Matrigel upon which 3-MA (10 mM) was added for 2 d. Representative fields of EtBr stained day 8 acini (middle) and corresponding phase contrast images (left) are shown. Bar, 20 μm. Right, day 8 acini were immunostained with α-cleaved caspase-3 (green) and α-laminin-5 (red). DAPI-stained equatorial confocal cross-sections are shown. Bar, 20 μm. (B) After 6 d of 3D culture, 3-MA (10 mM) was added to the indicated cell types; the percentage of acini containing EtBr-positive cells was enumerated on subsequent days. Results are the mean ± SEM of four independent experiments. (C) After 6 d of 3D culture, CQ (20 μM) or BafA (20 nM) was added to the indicated cell types; the percentage of acini containing EtBr-positive cells was enumerated on subsequent days. Results are the mean ± SEM of three independent experiments.
Effect of ATG5 or ATG7 Depletion on Luminal Apoptosis and Lumen Formation
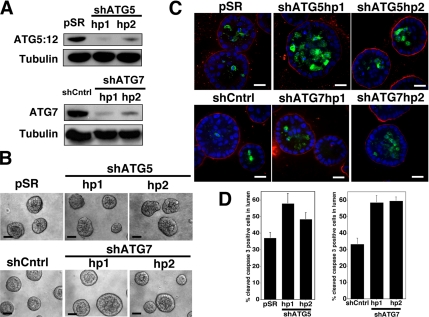
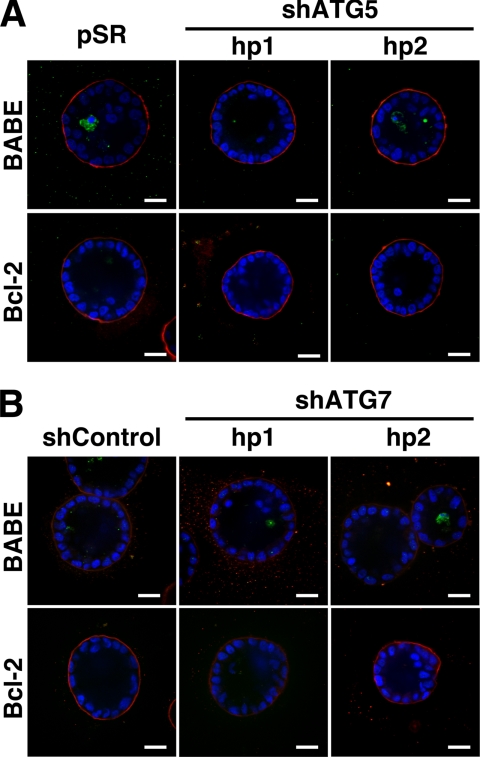
In addition, we interrogated how the stable depletion of ATG5 or ATG7 affected apoptosis and luminal filling in 3D culture. MCF-10A cells expressing two independent small hairpin RNAs against ATG5 (shATG5 hp 1 and 2) or ATG7 (shATG7 hp1 and 2) were generated. In cells expressing these hairpins, we confirmed the stable reduction of the ATG5:12 complex or ATG7 (Figure 8A). In these cells, we observed potent knockdown for >30 d, rendering them suitable for use in long-term 3D morphogenesis assays. Acini derived from shATG5- and shATG7-expressing cells formed polarized structures that morphologically resembled controls (Figure 8B). Furthermore, during lumen formation, we observed high levels of cleaved caspase-3 within these structures; in fact, the lumens of both ATG5 and ATG7 depleted acini contained increased cleaved caspase-3–positive cells compared with controls (Figure 8, C and D). Finally, to examine whether the combined reduction of autophagy and apoptosis would elicit the long-term survival of matrix-detached cells occupying the lumens of 3D acini, we generated cells with Bcl-2 overexpression plus stable ATG5 or ATG7 knockdown. However, we did not observe significant luminal filling in cells coexpressing Bcl2 + shATG5 (Figure 9A) or Bcl2 + shATG7 (Figure 9B) over 20 d of 3D culture. Similar to both wild-type and Bcl-2–expressing controls, these structures exhibited a hollow lumen. Thus, ATG5 or ATG7 depletion does not cooperate with Bcl-2 to fill the lumen.
Figure 8.
Effect of Stable ATG5 or ATG7 depletion on luminal apoptosis. (A) Top, MCF-10A cells were infected with retrovirus encoding an empty control vector (pSR) or two independent small hairpin RNAs to knockdown ATG5 (shATG5 hp1 and 2). Bottom, MCF-10A cells were infected with lentiviral particles expressing a nontargeting hairpin (shCntrl) or two independent small hairpin RNAs to knockdown ATG7 (shATG7 hp1 and 2). Lysates were immunoblotted with α-ATG7 and α-tubulin. (B) Representative phase contrast images of acini generated from control (pSR or shCntrl), shATG5 (hp 1 and 2), and shATG7 (hp1 and 2) cells after 18 d of 3D culture. Bar, 20 μm. (C) Indicated cell types grown in 3D for 12 d were immunostained for α-cleaved caspase-3 (green), α-laminin-5 (red), and counterstained with DAPI to detect nuclei (blue). Confocal cross sections of equators are shown. Bar, 20 μM. (D) Acini from the indicated cell types were fixed and immunostained on day 12 as illustrated in C. Cells occupying the lumen were defined as those lacking direct contact with basement membrane as delineated by laminin 5 staining. The percentage of cells positive for cleaved caspase-3 (mean ± SEM) was counted in 99 acini obtained from three independent 3D culture experiments.
Figure 9.
Effect of stable ATG5 or ATG7 depletion on lumen formation. (A and B) Day 20 acini from the indicated cell types grown were immunostained for α-cleaved caspase-3 (green), α-laminin-5 (red), and counterstained with DAPI to detect nuclei (blue). Confocal equatorial cross-sections are shown. Bar, 20 μm.
DISCUSSION
Here, we demonstrate that ECM detachment induces autophagy in various nontumorigenic epithelial cell lines and in primary human epithelial cells. We also discover that ATG depletion results in both enhanced cleaved caspase-3 during anoikis and reduced clonogenic viability upon reattachment. Remarkably, matrix-detached cells still exhibit autophagy when apoptosis is blocked by Bcl-2 expression, and ATG depletion impairs the clonogenic survival of detached Bcl-2–expressing cells. Thus, autophagy promotes epithelial cell survival during anoikis, including detached cells harboring antiapoptotic lesions.
Anoikis serves as an important mechanism to maintain tissue homeostasis by killing cells that have lost contact with an underlying basement membrane (Gilmore, 2005). However, evidence indicates that the detachment of nonmalignant cells also triggers antiantiapoptotic signals, such as nuclear factor-κB and inhibitor of apoptosis protein family members; these antiapoptotic mechanisms presumably delay the onset of apoptosis and allow cells to survive if they are able to reestablish ECM contact in a timely manner (Yan et al., 2005; Liu et al., 2006). Our results indicate, similar to these antiapoptotic pathways, the induction of autophagy also contributes to cell viability during ECM detachment.
Overall, our results resemble other circumstances where autophagy protects cells exposed to various forms of duress, including nutrient deprivation, growth factor withdrawal, ER stress, and ischemia (Boya et al., 2005; Lum et al., 2005; Degenhardt et al., 2006; Ogata et al., 2006; Kouroku et al., 2007). Autophagy may promote cell survival by a number of mechanisms, such as 1) generating nutrients and energy to sustain starving or stressed cells, 2) degrading toxic proteins, or 3) protecting cells from oxidative stress by sequestering damaged mitochondria (Debnath et al., 2005; Levine and Yuan, 2005). Further studies are required to identify the precise mechanisms through which detachment-induced autophagy promotes cell survival. Moreover, in addition to regulating survival, autophagy may direct other critical biological functions during matrix detachment; for example, autophagy suppresses both DNA damage and chromosomal instability in response to metabolic stress (Karantza-Wadsworth et al., 2007; Mathew et al., 2007).
Because integrin engagement is critical for the proper transduction of growth factor receptor mediated signals, we hypothesized that detachment-induced autophagy results from reduced growth factor receptor signaling. Importantly, in human mammary epithelial cells, ECM contact is required for both EGFR expression and the activation of critical downstream signals (Reginato et al., 2003). However, we demonstrate that even though enforced EGFR expression during anoikis can restore certain downstream survival-promoting signals, such as ERK activation, it is not sufficient to prevent detachment-induced autophagy. Thus, we postulate that other cell adhesion regulated stress pathways also direct detachment-induced autophagy. We have identified multiple signals during ECM detachment that may positively regulate autophagy, including AMPK activation and the phosphorylation of eIF2α; in ongoing studies, we are trying to dissect how these signals, either individually or in combination, contribute to detachment-induced autophagy.
Remarkably, a recent study demonstrates that integrin engagement, most notably α3β1, is actually required to sustain autophagy in starved prostate epithelial cells (Edick et al., 2007). In this study, the observed reductions in autophagy were based exclusively on measurements of GFP-LC3 puncta; thus, it remains unknown whether the reductions in punctate GFP-LC3 observed upon integrin blockade are actually due to decreased autophagosome formation versus increased LC3-II turnover in the lysosome (Tanida et al., 2005). Alternatively, detachment-induced autophagy may be cell type or context dependent. Importantly, whereas human mammary epithelial cells die after 24–48 h of detachment, certain epithelial cells, notably primary mouse mammary epithelial cells and rat intestinal epithelial cells, perish within a few hours following substratum detachment; thus, one can speculate that detachment-induced autophagy will not contribute to the viability of cells that undergo rapid anoikis (Gilmore, 2005).
Protection from anoikis is thought to promote filling of the normally hollow lumen in glandular epithelial structures, a hallmark of early epithelial cancers, such as carcinomas in situ (Debnath and Brugge, 2005; Gilmore, 2005). Although our previous work suggested that cells undergo autophagy as they die during 3D morphogenesis, two results in this study argue against a direct role for type 2 death during lumen formation. First, the acute pharmacological inhibition of autophagy during morphogenesis enhances luminal cell death. Second, the knockdown of ATG5 or ATG7, two proteins critical for early autophagosome formation, enhances luminal apoptosis during MCF-10A 3D morphogenesis and fails to elicit long-term luminal filling, even when combined with apoptosis inhibition. Our results are consistent with recent studies in E1A immortalized mouse mammary cells lacking one allele of Beclin; when grown in 3D culture, cells with reduced Beclin exhibit increased lumen formation compared with wild-type controls (Karantza-Wadsworth et al., 2007). Interestingly, in embryoid bodies derived from cells lacking atg5 or beclin1, apoptotic cell corpses fail to clear during cavitation, because autophagy is critical for the presentation of signals, such as phosphatidylserine, that mediate the phagocytic clearance of apoptotic cells (Qu et al., 2007). Accordingly, the increased numbers of apoptotic cells that we observe when autophagy is inhibited during lumen formation could arise from defective corpse clearance. However, over longer times, we do not observe reduced clearance in ATG5 or ATG7-depleted structures; rather, these structures form hollow lumens at rates similar to controls. Clearance may ultimately proceed during 3D morphogenesis because we have only reduced these ATGs rather than completely eliminated these proteins in acini. Nonetheless, recent work indicates that inhibition of autophagy, a late stage event observed in MCF7 carcinoma cells during anoikis, does not prevent phagocytic engulfment (Petrovski et al., 2007). Importantly, during both 3D lumen formation and ECM detachment, we have observed the robust induction of autophagy in Bcl-2–expressing cells, indicating that autophagy can proceed independently of apoptosis in ECM-detached cells, rather than merely serve as a secondary clearance mechanism to remove cells undergoing apoptosis (Debnath et al., 2002). Further delineating the respective roles of autophagy in luminal cell survival versus the phagocytic clearance of dying cells during lumen formation remains a topic for future investigation.
Several autophagy regulators are down-regulated in human cancers, including BECN-1/ATG6, which is monoallelically deleted in 40–75% of breast, prostate, and ovarian tumors, and Death Associated Protein Kinase 1, which is methylated in many tumors (Liang et al., 1999; Inbal et al., 2002). The importance of autophagy as a tumor suppressor is further supported by the development of tumors in becn-1+/− mice (Qu et al., 2003; Yue et al., 2003). In contrast, because autophagy has well-established cytoprotective functions, it can promote the survival of tumor cells exposed to stresses such as hypoxia, nutrient limitation, and chemotherapy (Ogier-Denis and Codogno, 2003; Jin et al., 2007). Both of these opposing functions may be relevant to cancer progression and treatment. Our results broach the possibility that autophagy contributes to the survival of oncogenic cells lacking appropriate matrix contact. Indeed, the ability to survive in the absence of normal ECM is considered a critical feature of metastasis, because cancer cells in the bloodstream or secondary tissue sites are either deprived of matrix or exposed to foreign matrix components (Chambers et al., 2002). Accordingly, we are currently investigating how oncogenic pathways regulate autophagy during ECM detachment and determining whether autophagy contributes to the survival and expansion of oncogene-expressing cells undergoing anoikis and in the lumens of 3D structures.
Supplementary Material
ACKNOWLEDGMENTS
We are especially grateful to Dr. Joan Brugge (Harvard Medical School), in whose laboratory this work was initiated. Drs. Noburu Mizushima and Tamotsu Yoshimori generously provided reagents and cells. Grant support includes National Institutes of Health KO8 Award (CA-098419), Culpeper Scholar Award (Partnership for Cures), an American Association for Cancer Research/Genentech BioOncology Career Award, a Howard Hughes Medical Institute Early Career Award, and funds from the UCSF Sandler Program in Basic Sciences (all to J.D.).
Abbreviations used:
- ATG
autophagy gene
- AMPK
AMP-activated protein kinase
- ECM
extracellular matrix
- EGFR
epidermal growth factor receptor
- eIF2α
eukaryotic initiation factor 2α
- GFP-LC3
green fluorescent protein fused to light chain 3
- LC3
light chain 3
- PE
phosphotidylethanolamine
- 3D
three-dimensional.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-10-1092) on December 19, 2007.
REFERENCES
- Boya P., et al. Inhibition of macroautophagy triggers apoptosis. Mol. Cell Biol. 2005;25:1025–1040. doi: 10.1128/MCB.25.3.1025-1040.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers A. F., Groom A. C., MacDonald I. C. Dissemination and growth of cancer cells in metastatic sites. Nat. Rev. Cancer. 2002;2:563–572. doi: 10.1038/nrc865. [DOI] [PubMed] [Google Scholar]
- Debnath J., Baehrecke E. H., Kroemer G. Does autophagy contribute to cell death? Autophagy. 2005;1:e10–e18. doi: 10.4161/auto.1.2.1738. [DOI] [PubMed] [Google Scholar]
- Debnath J., Brugge J. S. Modelling glandular epithelial cancers in three-dimensional cultures. Nat. Rev. Cancer. 2005;5:675–688. doi: 10.1038/nrc1695. [DOI] [PubMed] [Google Scholar]
- Debnath J., Mills K. R., Collins N. L., Reginato M. J., Muthuswamy S. K., Brugge J. S. The role of apoptosis in creating and maintaining luminal space within normal and oncogene-expressing mammary acini. Cell. 2002;111:29–40. doi: 10.1016/s0092-8674(02)01001-2. [DOI] [PubMed] [Google Scholar]
- Debnath J., Muthuswamy S. K., Brugge J. S. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 2003;30:256–268. doi: 10.1016/s1046-2023(03)00032-x. [DOI] [PubMed] [Google Scholar]
- Degenhardt K., et al. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell. 2006;10:51–64. doi: 10.1016/j.ccr.2006.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edick M. J., Tesfay L., Lamb L. E., Knudsen B. S., Miranti C. K. Inhibition of integrin-mediated crosstalk with EGFR/Erk or Src signaling pathways in autophagic prostate epithelial cells induces caspase-independent death. Mol. Biol. Cell. 2007;18:2481–2490. doi: 10.1091/mbc.E06-04-0261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisch S. M. Methods for studying anoikis. Methods Mol. Biol. 1999;129:251–256. doi: 10.1385/1-59259-249-X:251. [DOI] [PubMed] [Google Scholar]
- Frisch S. M., Francis H. Disruption of epithelial cell-matrix interactions induces apoptosis. J. Cell Biol. 1994;124:619–626. doi: 10.1083/jcb.124.4.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmore A. P. Anoikis. Cell Death Differ. 2005;12(suppl 2):1473–1477. doi: 10.1038/sj.cdd.4401723. [DOI] [PubMed] [Google Scholar]
- Hoyer-Hansen M., et al. Control of macroautophagy by calcium, calmodulin-dependent kinase kinase-beta, and Bcl-2. Mol. Cell. 2007;25:193–205. doi: 10.1016/j.molcel.2006.12.009. [DOI] [PubMed] [Google Scholar]
- Inbal B., Bialik S., Sabanay I., Shani G., Kimchi A. DAP kinase and DRP-1 mediate membrane blebbing and the formation of autophagic vesicles during programmed cell death. J. Cell Biol. 2002;157:455–468. doi: 10.1083/jcb.200109094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin S., DiPaola R. S., Mathew R., White E. Metabolic catastrophe as a means to cancer cell death. J. Cell Sci. 2007;120:379–383. doi: 10.1242/jcs.03349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y., Mizushima N., Ueno T., Yamamoto A., Kirisako T., Noda T., Kominami E., Ohsumi Y., Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karantza-Wadsworth V., Patel S., Kravchuk O., Chen G., Mathew R., Jin S., White E. Autophagy mitigates metabolic stress and genome damage in mammary tumorigenesis. Genes Dev. 2007;21:1621–1635. doi: 10.1101/gad.1565707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsumi A., Orr A. W., Tzima E., Schwartz M. A. Integrins in mechanotransduction. J. Biol. Chem. 2004;279:12001–12004. doi: 10.1074/jbc.R300038200. [DOI] [PubMed] [Google Scholar]
- Klionsky D. J., et al. A unified nomenclature for yeast autophagy-related genes. Dev. Cell. 2003;5:539–545. doi: 10.1016/s1534-5807(03)00296-x. [DOI] [PubMed] [Google Scholar]
- Kouroku Y., Fujita E., Tanida I., Ueno T., Isoai A., Kumagai H., Ogawa S., Kaufman R. J., Kominami E., Momoi T. ER stress (PERK/eIF2alpha phosphorylation) mediates the polyglutamine-induced LC3 conversion, an essential step for autophagy formation. Cell Death Differ. 2007;14:230–239. doi: 10.1038/sj.cdd.4401984. [DOI] [PubMed] [Google Scholar]
- Kuma A., Hatano M., Matsui M., Yamamoto A., Nakaya H., Yoshimori T., Ohsumi Y., Tokuhisa T., Mizushima N. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- Kuma A., Matsui M., Mizushima N. LC3, an autophagosome marker, can be incorporated into protein aggregates independent of autophagy: caution in the interpretation of LC3 localization. Autophagy. 2007;3:323–328. doi: 10.4161/auto.4012. [DOI] [PubMed] [Google Scholar]
- Lee C. Y., Baehrecke E. H. Steroid regulation of autophagic programmed cell death during development. Development. 2001;128:1443–1455. doi: 10.1242/dev.128.8.1443. [DOI] [PubMed] [Google Scholar]
- Levine B., Klionsky D. J. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev. Cell. 2004;6:463–477. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- Levine B., Yuan J. Autophagy in cell death: an innocent convict? J. Clin. Invest. 2005;115:2679–2688. doi: 10.1172/JCI26390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J., et al. The energy sensing LKB1-AMPK pathway regulates p27(kip1) phosphorylation mediating the decision to enter autophagy or apoptosis. Nat. Cell Biol. 2007;9:218–224. doi: 10.1038/ncb1537. [DOI] [PubMed] [Google Scholar]
- Liang X. H., Jackson S., Seaman M., Brown K., Kempkes B., Hibshoosh H., Levine B. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–676. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- Liu Z., et al. Detachment-induced upregulation of XIAP and cIAP2 delays anoikis of intestinal epithelial cells. Oncogene. 2006;25:7680–7690. doi: 10.1038/sj.onc.1209753. [DOI] [PubMed] [Google Scholar]
- Lum J. J., Bauer D. E., Kong M., Harris M. H., Li C., Lindsten T., Thompson C. B. Growth factor regulation of autophagy and cell survival in the absence of apoptosis. Cell. 2005;120:237–248. doi: 10.1016/j.cell.2004.11.046. [DOI] [PubMed] [Google Scholar]
- Martin D. N., Baehrecke E. H. Caspases function in autophagic programmed cell death in Drosophila. Development. 2004;131:275–284. doi: 10.1242/dev.00933. [DOI] [PubMed] [Google Scholar]
- Mathew R., Kongara S., Beaudoin B., Karp C. M., Bray K., Degenhardt K., Chen G., Jin S., White E. Autophagy suppresses tumor progression by limiting chromosomal instability. Genes Dev. 2007;21:1367–1381. doi: 10.1101/gad.1545107. [correction published in Genes Dev. (2007) 21, 1701] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meley D., Bauvy C., Houben-Weerts J. H., Dubbelhuis P. F., Helmond M. T., Codogno P., Meijer A. J. AMP-activated protein kinase and the regulation of autophagic proteolysis. J. Biol. Chem. 2006;281:34870–34879. doi: 10.1074/jbc.M605488200. [DOI] [PubMed] [Google Scholar]
- Meredith J. E., Jr, Fazeli B., Schwartz M. A. The extracellular matrix as a cell survival factor. Mol. Biol. Cell. 1993;4:953–961. doi: 10.1091/mbc.4.9.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills K. R., Reginato M., Debnath J., Queenan B., Brugge J. S. Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) is required for induction of autophagy during lumen formation in vitro. Proc. Natl. Acad. Sci. USA. 2004;101:3438–3443. doi: 10.1073/pnas.0400443101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranti C. K., Brugge J. S. Sensing the environment: a historical perspective on integrin signal transduction. Nat. Cell Biol. 2002;4:E83–E90. doi: 10.1038/ncb0402-e83. [DOI] [PubMed] [Google Scholar]
- Ogata M., et al. Autophagy is activated for cell survival after endoplasmic reticulum stress. Mol. Cell Biol. 2006;26:9220–9231. doi: 10.1128/MCB.01453-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogier-Denis E., Codogno P. Autophagy: a barrier or an adaptive response to cancer. Biochim. Biophys. Acta. 2003;1603:113–128. doi: 10.1016/s0304-419x(03)00004-0. [DOI] [PubMed] [Google Scholar]
- Pattingre S., Tassa A., Qu X., Garuti R., Liang X. H., Mizushima N., Packer M., Schneider M. D., Levine B. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122:927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- Petrovski G., Zahuczky G., Katona K., Vereb G., Martinet W., Nemes Z., Bursch W., Fesus L. Clearance of dying autophagic cells of different origin by professional and non-professional phagocytes. Cell Death Differ. 2007;14:1117–1128. doi: 10.1038/sj.cdd.4402112. [DOI] [PubMed] [Google Scholar]
- Qu X., et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J. Clin. Invest. 2003;112:1809–1820. doi: 10.1172/JCI20039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu X., Zou Z., Sun Q., Luby-Phelps K., Cheng P., Hogan R. N., Gilpin C., Levine B. Autophagy gene-dependent clearance of apoptotic cells during embryonic development. Cell. 2007;128:931–946. doi: 10.1016/j.cell.2006.12.044. [DOI] [PubMed] [Google Scholar]
- Reginato M. J., Mills K. R., Paulus J. K., Lynch D. K., Sgroi D. C., Debnath J., Muthuswamy S. K., Brugge J. S. Integrins and EGFR coordinately regulate the pro-apoptotic protein Bim to prevent anoikis. Nat. Cell Biol. 2003;5:733–740. doi: 10.1038/ncb1026. [DOI] [PubMed] [Google Scholar]
- Rytomaa M., Lehmann K., Downward J. Matrix detachment induces caspase-dependent cytochrome c release from mitochondria: inhibition by PKB/Akt but not Raf signalling. Oncogene. 2000;19:4461–4468. doi: 10.1038/sj.onc.1203805. [DOI] [PubMed] [Google Scholar]
- Shimizu S., Kanaseki T., Mizushima N., Mizuta T., Arakawa-Kobayashi S., Thompson C. B., Tsujimoto Y. Role of Bcl-2 family proteins in a non-apoptotic programmed cell death dependent on autophagy genes. Nat. Cell Biol. 2004;6:1221–1228. doi: 10.1038/ncb1192. [DOI] [PubMed] [Google Scholar]
- Talloczy Z., Jiang W., Virgin H.W.T., Leib D. A., Scheuner D., Kaufman R. J., Eskelinen E. L., Levine B. Regulation of starvation- and virus-induced autophagy by the eIF2alpha kinase signaling pathway. Proc. Natl. Acad. Sci. USA. 2002;99:190–195. doi: 10.1073/pnas.012485299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanida I., Minematsu-Ikeguchi N., Ueno T., Kominami E. Lysosomal turnover, but not a cellular level, of endogenous LC3 is a marker for autophagy. Autophagy. 2005;1:84–91. doi: 10.4161/auto.1.2.1697. [DOI] [PubMed] [Google Scholar]
- Underwood J. M., Imbalzano K. M., Weaver V. M., Fischer A. H., Imbalzano A. N., Nickerson J. A. The ultrastructure of MCF-10A acini. J. Cell Physiol. 2006;208:141–148. doi: 10.1002/jcp.20639. [DOI] [PubMed] [Google Scholar]
- Yan S. R., Joseph R. R., Rosen K., Reginato M. J., Jackson A., Allaire N., Brugge J. S., Jobin C., Stadnyk A. W. Activation of NF-kappaB following detachment delays apoptosis in intestinal epithelial cells. Oncogene. 2005;24:6482–6491. doi: 10.1038/sj.onc.1208810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L., Alva A., Su H., Dutt P., Freundt E., Welsh S., Baehrecke E. H., Lenardo M. J. Regulation of an ATG7-beclin 1 program of autophagic cell death by caspase-8. Science. 2004;304:1500–1502. doi: 10.1126/science.1096645. [DOI] [PubMed] [Google Scholar]
- Yu L., Wan F., Dutta S., Welsh S., Liu Z., Freundt E., Baehrecke E. H., Lenardo M. Autophagic programmed cell death by selective catalase degradation. Proc. Natl. Acad. Sci. USA. 2006;103:4952–4957. doi: 10.1073/pnas.0511288103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue Z., Jin S., Yang C., Levine A. J., Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc. Natl. Acad. Sci. USA. 2003;100:15077–15082. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.