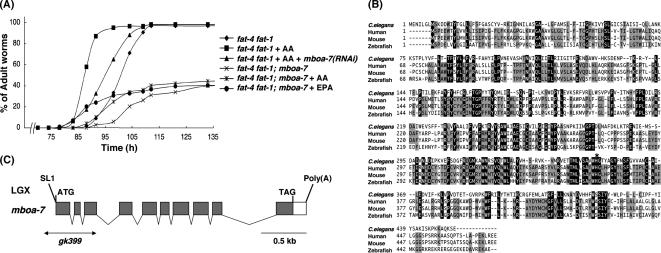
Figure 2.
(A) Knockdown and knockout of mboa-7 inhibit growth rescue by dietary supplementation with AA or EPA. Growth of each strain was scored as described in Figure 1B, except that E. coli HT115 was used as a food source. All experiments were performed at 15°C. (B) Multiple sequence alignment of C. elegans mboa-7 and homologous sequences in human (BB1/LENG4; Fukunaga-Johnson et al., 1996; Wende et al., 2000), mouse, and zebrafish. Residues identical in all four sequences are shaded in black, and residues identical in three proteins are shaded in gray. The numbers on the left indicate amino acid positions. The histidine residue indicated by asterisk is the predicted active site of the MBOAT motif in mboa-7 (Hofmann, 2000; Bosson et al., 2006). Accession numbers for the sequences used were as follows: C. elegans, EU016382; human, EU016381; mouse, EU016380; and zebrafish, NP_609029. (C) Genomic structure of mboa-7 (F14F3.3). The mboa-7 gene is located on chromosome X. Gray boxes indicate coding exons, and white boxes indicate 5′ and 3′ untranslated sequences. The positions of the ATG initiation codon, stop codon (TAG), the trans-spliced leader SL1, and the poly(A) tail are shown. The extent of the deletion in mboa-7(gk399) is indicated by a horizontal line.