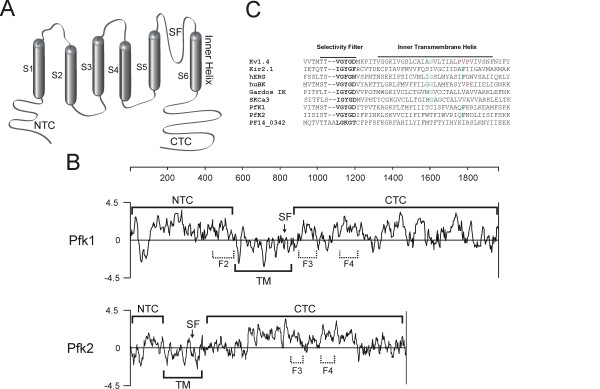
Figure 1.
PfK1 and PfK2. A. Membrane topology schematic of a single K+ channel subunit. Predicted topographical domains are indicated: NTC = N-terminal cytoplasmic portion; CTC = C-terminal cytoplasmic portion; SF = selectivity filter; and S1–S6 = membrane spanning domains. B. Hydropathy plots of the predicted protein sequences of PfK1 and PfK2. Negative deflections signify hydrophobic sequences. NTC = N-terminal cytoplasmic portion; CTC = C-terminal cytoplasmic portion; TM = Transmembrane portion and SF = selectivity filter. F2, F3 and F4 refer to protein segments used to generate specific antisera. C. Alignment of channel conduction pore protein sequences of various well characterized human K+ channels, highlighting features common to the family (G = suggested flexibility point for voltage-gated K+ channels. P = possible bend-inducing prolines. F = sites for possible drug block). Kv1.4 = voltage-gated Shaker-type K+ channel, Kir2.1 = inward rectifying K+ channel, hERG = human Ether a-gogo-Related Gene channel, huBK = human BK Ca2+-activated K+ channel, Gardos IK K+ channel and SKCa3 SK Ca2+-activated K+ channel. The sequence encoded by PF14_0342 is also included in the alignment, due to its suggestion as being a possible third parasite-encoded K+ channel [1]. PF14_0342 does not have several features common to K+ channels, including the pore region GYG motif.