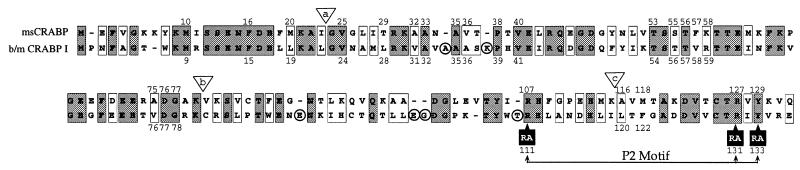
Figure 4.
Alignment of the msCRABP and b/m CRABP I amino acid sequences. Identities and conservative substitutions are shown with shaded and open boxes, respectively. Residues postulated to form the ligand binding pocket (see Fig. 5B) based on proximity to bound RA (defined as those residues with at least one heavy atom located within 5.5 Å of any ligand heavy atom) in b/m CRABP I are numbered. Three highly conserved residues (P2 Motif) considered essential for binding RA are indicated by solid boxes. Residues deleted in the molecular modeling are circled. The b/m CRABP I residue numbering is that used for the crystal structure analysis (15). B/m amino acid sequences are identical. The positions of the three introns are shown with lettered inverted triangles.