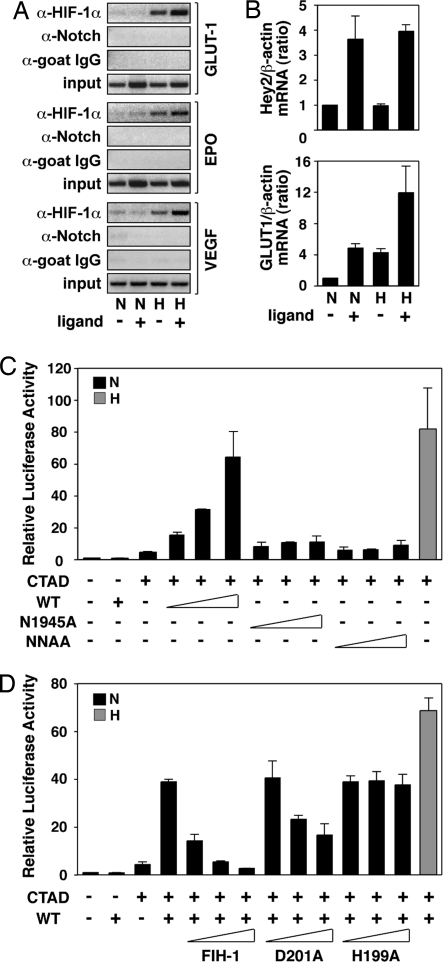
Fig. 5.
Activation of Notch increases binding of HIF-1α to target genes and causes derepression of HIF-1α function. (A) Binding of HIF-1α to target gene HREs by ChIP analysis in the absence or presence of activated Notch. C2C12 cells stably expressing full-length Notch were cultured in the presence of control 293T cells (ligand −) or 293T cells stably expressing Notch ligand Serrate-1 (ligand +) and kept at normoxia (N) or exposed to hypoxia (H). ChIP assays were performed using the indicated antibodies, and RT-PCR analysis of target gene HREs was carried out. (B) Activation of the Notch pathway increases HIF-1 target gene expression. 293T cells stably expressing full-length Notch were cultured on plates coated with IgG (ligand −) or Jagged-1 (ligand +) and kept at normoxia (N) or hypoxia (H). Total RNA was prepared, and quantitative RT-PCR was performed. (C) Notch ICD derepresses HIF-1α CAD function. P19 cells were transfected with 0.5 μg of a GAL4-driven luciferase plasmid and 50 ng of GAL-fused HIF-1α C-TAD expression plasmid in the presence of increasing amounts (50, 100, and 200 ng) of plasmids encoding wild-type or Asn mutant Notch1 ICD. (D) Expression of wild-type FIH-1 restores repression of CAD function at normoxia. P19 cells were transfected with 0.5 μg of a GAL4-driven luciferase plasmid together with a GAL-fused HIF-1α CAD expression plasmid (50 ng) and a plasmid encoding wild-type Notch 1 ICD (200 ng) in the presence of increasing amounts (50, 100, 200 ng) of plasmids encoding wild-type or mutant (D201A and D199A) FIH-1. Data are presented as luciferase activity relative to cells transfected with GAL4-Luc. Values represent the mean ± S.E. of three independent experiments performed in duplicate (C and D).