Fig. 1.
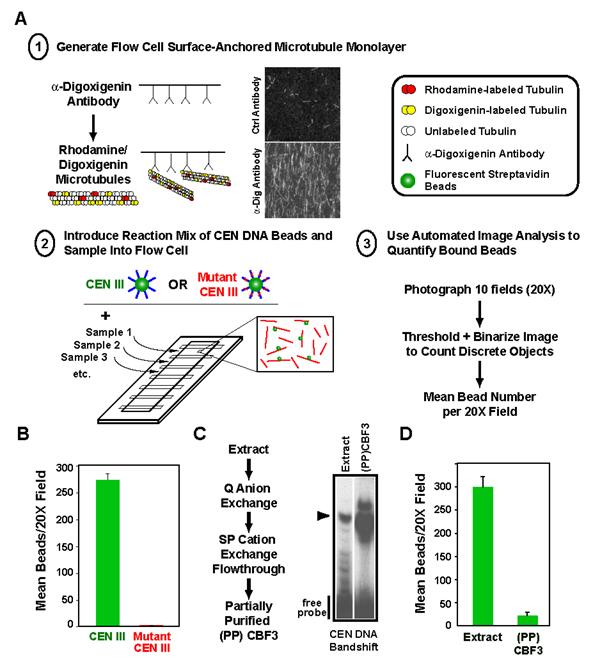
A quantitative in vitro assay for the binding of budding yeast CEN DNA to microtubules. (A) Schematic of the in vitro assay. Adaptations to the original scheme important for quantitative biochemical analysis are emphasized here and include: 1) stable adsorption of microtubules using tubulin covalently modified with digoxigenin; 2) multiplexing of flow cells on a single slide; and 3) automated image analysis to measure number of bound beads. For quantitation, 10 fields at 20X magnification are photographed per sample and averaged. (B) Linkage of beads to microtubules is observed with wild-type but not mutant CEN DNA. The mutant harbors a deletion of the central CCG in CDEIII that prevents binding of the CBF3 complex and abolishes centromere activity in vivo. Error bars=SD. (C) Partial purification of CBF3 using a CEN DNA gel-shift assay. The flowchart describes the chromatography steps and the gel panel shows enrichment of the CEN DNA bandshift relative to starting extract in the partially purified (PP) fraction. The arrowhead marks the CEN DNA-CBF3 complex. (D) Partially purified CBF3 does not link CEN DNA beads to microtubules. Note that the volume of starting extract used to prepare the CBF3 added to the (PP)CBF3 reaction is ∼25-fold greater than that assayed in the extract reaction. If equivalent extract volumes are assayed, no binding is observed with (PP)CBF3. Error bars=SD.
