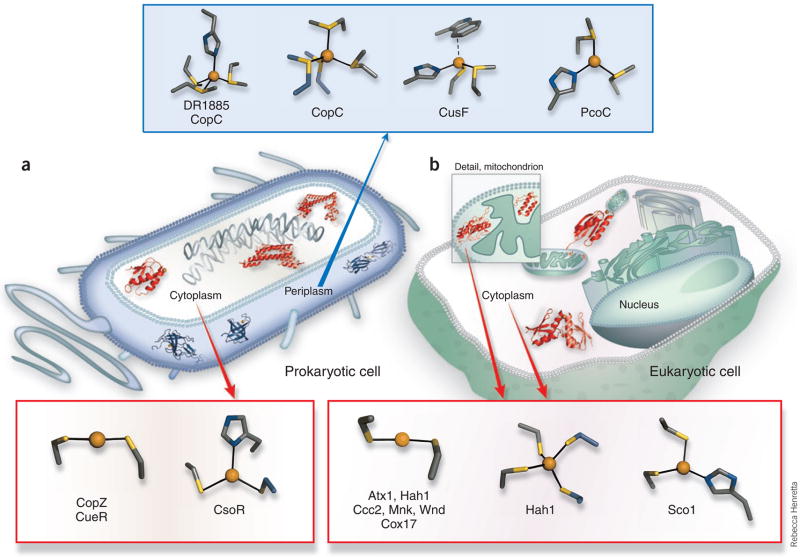
Figure 2.
Characterized Cu(I) trafficking and sensing coordination sites classified by cellular localization and function. (a) Protein structures depicted in theprokaryotic cell are the periplasmic putative trafficking proteins, which all form positively charged Cu(I) complexes (colored blue): Cu(I)-DR1885 (ref. 25),Cu(I)-CusF26 and Cu(I)2CopC224); and the cytoplasmic proteins, which all form negatively charged Cu(I) complexes (colored red): the chaperone Cu(I)-CopZ11and the metalloregulatory DNA-binding proteins Cu(I)2CsoR2 (ref. 15) and Cu(I)2CueR2 (ref. 6). (b) In the cytosol of the eukaryotic cell (not drawn to scale),two chaperone structures are shown, the yeast Cu(I)-Atx1 (ref. 7) and the human Cu(I)-Hah12 (ref. 12). A number of mitochondrial Cco assembly proteinshave suspected Cu(I) transport function, and the Cu(I)-Sco1 (ref. 17) and Cu(I)-Cox17 (ref. 20) structures are shown. The coordination sites are shown indetail outside of the cell diagrams, and have been characterized by a combination of crystallographic, XAS and NMR structural studies. In addition to theprotein sites listed above, a Met3His site of CopC23 and a Met2His site of PcoC22 are included with the methionine-rich cationic sites of the prokaryoticperiplasmic proteins. The anionic Cys2Cu(I) sites of the eukaryotic cytosolic proteins Hah1 (ref. 30) and Atx1 (ref. 7) and the cytosolic metal-bindingdomains of the Ccc2, Menkes (Mnk)9 and Wilson (Wnd)10 disease Cu(I) transporting P-type ATPases are also included.