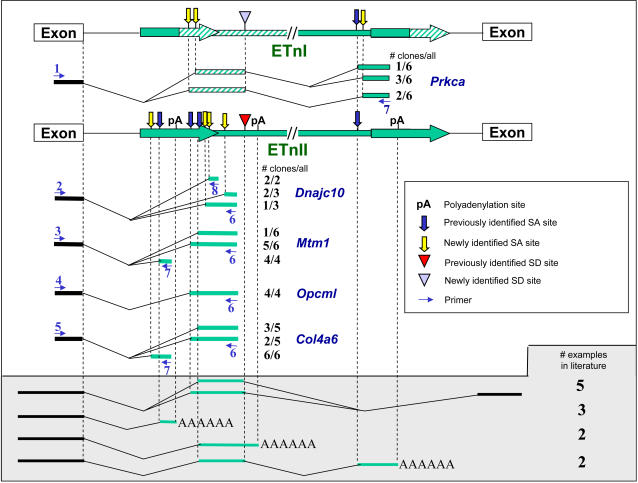
Figure 5. Detection of ETn-gene chimeric transcripts.
Aberrant transcripts induced by a novel ETn insertion into a gene intron are shown. Gene direction, from left to right, is the same as ETn orientation. Genes Dnajc10, Opcml, Mtm1 and Col4a6 harbor ETnII insertions, while Prkca has an ETnI insertion. ETn sections that are different in an ETnI element compared to ETnII are shown as striped. Cryptic and natural splice acceptor sites are designated as blue (previously identified) or yellow (newly identified in this study) vertical arrows. Splice donor sites are represented as red (previously known) or light blue (newly identified) triangles. Natural and cryptic polyadenylation sites are marked as pA. Numbered thin arrows denote primers used to amplify chimeric transcripts. Sense primers in the upstream exon are as follows: 1, Prkca-up-ex-s; 2, Dnajc10-up-ex-s; 3, Mtm1-up-ex-s; 4, Opcml-up-ex-s; 5, Col4a6-up-ex-s. Antisense primers in the ETn are as follows: 6, IM_3as; 7, MusD2_7130as; 8, LTR_2as. Number of clones for each transcript variant compared to all clones sequenced for each primer pair is indicated. Chimeric ETn transcripts and number of their examples identified previously [8] are shown at the bottom. Sequences of all splice sites used in these cases are shown in Figure S4.