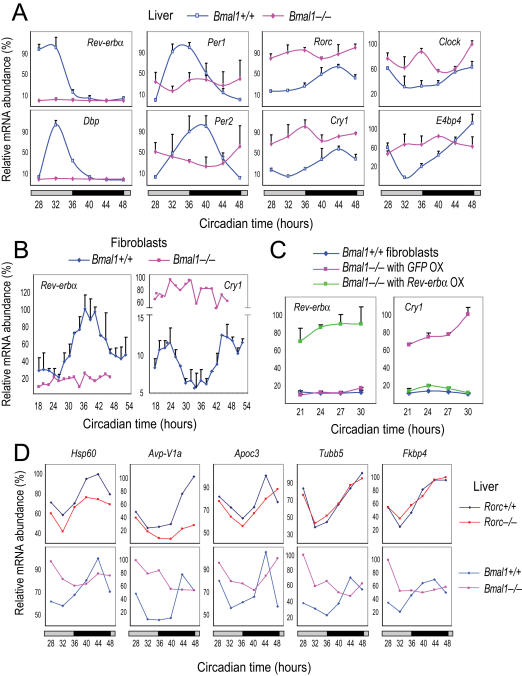
Figure 4. REV-ERBs play a prominent role in combinatorial regulation of Cry1 and Rorc.
(A) Temporal mRNA expression profiles of clock genes in the liver of Bmal1−/− mice. Expression was analyzed at 4-hr intervals by Q-PCR. Values are expressed as percentage of maximum expression for each gene. Error bar represents standard deviation (SD) of expression levels from four mice. The clock genes are presented in four groups based on different mRNA expression patterns (phase and level) in WT and Bmal1−/− mice. For instance, transcription of Cry1 and Rorc is elevated, rather than repressed, in the Bmal1−/− liver. Circadian time: hours after animal release in constant darkness. (B) Temporal mRNA expression profiles of Rev-erbα and Cry1 in Bmal1−/− fibroblasts. Expression was analyzed at 2-hr intervals by Q-PCR. Values are expressed as percentage of maximum expression for each gene. Results were confirmed in two independent time courses. Error bars represent SD of two culture samples for each cell line. Cry1 mRNA levels are constantly high throughout the day and Rev-erbα expression is completely abolished in Bmal1−/− fibroblasts, similar to results obtained from the liver. Circadian time: hours after serum treatment. (C) Over-expression (OX) of Rev-erbα represses elevated Cry1 mRNA levels in Bmal1−/− fibroblasts. Expression of GFP and REV-ERBα is driven by a constitutive CAG promoter. Temporal mRNA expression was analyzed at 3-hr intervals by Q-PCR. Values are expressed as percentage of maximum expression for each gene. Results were confirmed in two independent experiments. Error bars represent SD of two culture samples for each cell line. REV-ERBα expression was confirmed by Q-PCR, and also by Western blotting (data not shown). Circadian time: hours after serum treatment. (D) Temporal mRNA expression profiles of clock-controlled output genes in the liver of Rorc −/− and Bmal1−/− mice. Experiments were performed as described in Figure 1A for Rorc −/− mice and Figure 4A for Bmal1−/− mice. As for Bmal1 and Cry1, the prominent role of REV-ERBs in regulating transcription explains the elevated mRNA levels of these output genes in Bmal1−/− mice. For clarity, error bars representing SD from four mice (<10% for each gene) were omitted. Circadian time: hours after animal release in constant darkness.