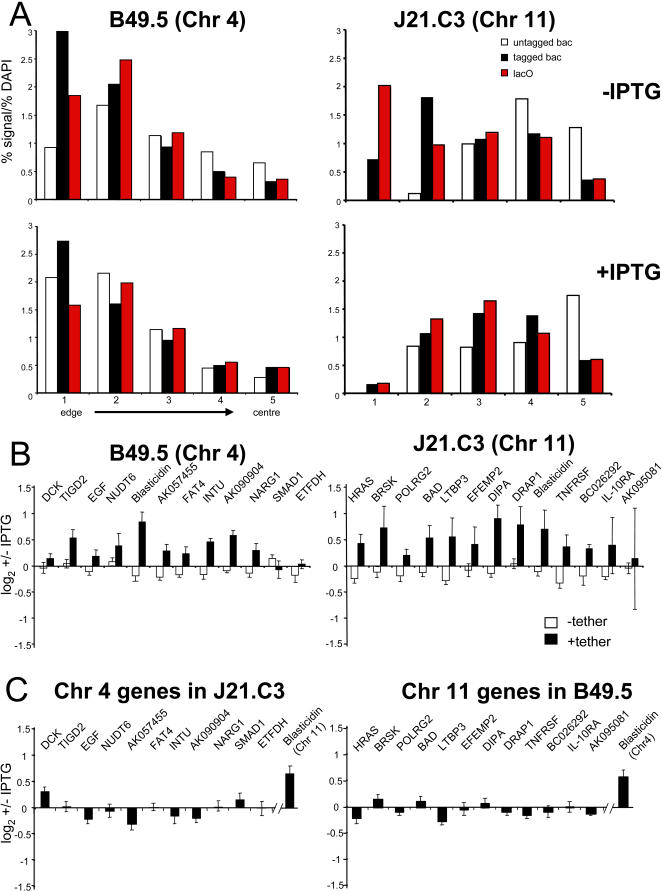
Figure 5. Reversibility of lacO-lacI Tethering and Gene Suppression.
(A) Histograms showing the mean proportion (%) of probe hybridisation signal, normalised to the proportion of DAPI stain (y axis), across the 5 concentric shells eroded from the periphery (shell 1) through to the centre (shell 5) of the nucleus (x axis), for a proximal BAC on the untagged chromosome (open bars) and on the tagged chromosome (black bars) and the lacO sites (red bars) in the lacI-lap2β expressing cell lines grown for 48 hours with (+) and without (−) IPTG. n = 50 for each cell line. (B) qRT-PCR analysis of expression from endogenous genes on chromosome 4 in B49.5 cells (left) or on chromosome11 in J21.C3 cells (right), as well as the blasticidin gene, in cells grown with (+) and without (−) IPTG. Graphs show the mean (+/−s.e.m) log2 +/− IPTG ratios from independent analyses normalised to β-actin. Black bars; data from cells expressing the lacI-lap2β tether, white bars; parental cells with no tethering protein. (C) qRT-PCR analysis of genes on chromosome 4 in J21.C3 cells (left) or genes on chromosome11 in B49.5 cells (right) expressing the lacI-lap2β tethering protein and treated with IPTG.