Figure 5.
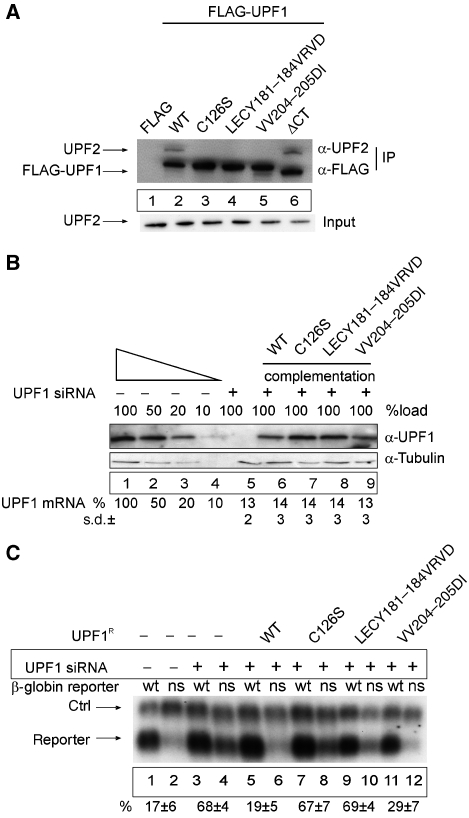
An UPF1 mutant that triggers NMD but fails to interact with UPF2. (A) Mutational analysis of the UPF2-binding site within the UPF1 CHR region. HeLa cells were transfected with empty FLAG plasmid (lane 1), wild-type FLAG-UPF1 (lane 2) or with the indicated FLAG-tagged mutants of UPF1 (lanes 3–6). Cell extract preparation, immunoprecipitation procedure and immunoblotting were carried out as described in Figure 3B in the presence of RNaseA. The UPF2 protein was detected by immunoblotting with an anti-UPF2 antibody. Lysate (5%) used for the immunoprecipitations was loaded in the input lane. (B, C) Functional analysis of UPF1 CHR mutants. (B) Functional complementation of UPF1 depletion. Immunoblotting analysis of lysates obtained from UPF1-depleted and UPF1-complemented cells was performed as described in Figure 2B and as described previously (Gehring et al, 2005). For the complementation of UPF1-depleted cells, cells were transfected with 0.2 μg FLAG-UPF1R. UPF1 mRNA levels were determined by qRT–PCR using primers that are specific for the endogenous mRNA and do not amplify the plasmid-derived UPF1 mRNA. The values were calculated from four independent experiments with standard deviations (±s.d.). (C) Northern blot analysis of RNA isolated from HeLa cells transfected with siRNA against β-galactosidase (lanes 1–2) or UPF1 (lanes 3–12). At 48 h after siRNA transfections, the cells were co-transfected with plasmids for the transfection efficiency control (ctrl), the NMD reporters (reporter; wt—wild-type β-globin, ns—NS39 β-globin mutant) and plasmids expressing the depicted FLAG-UPF1R variants. The values were calculated from three independent experiments. Error bars represent s.d.