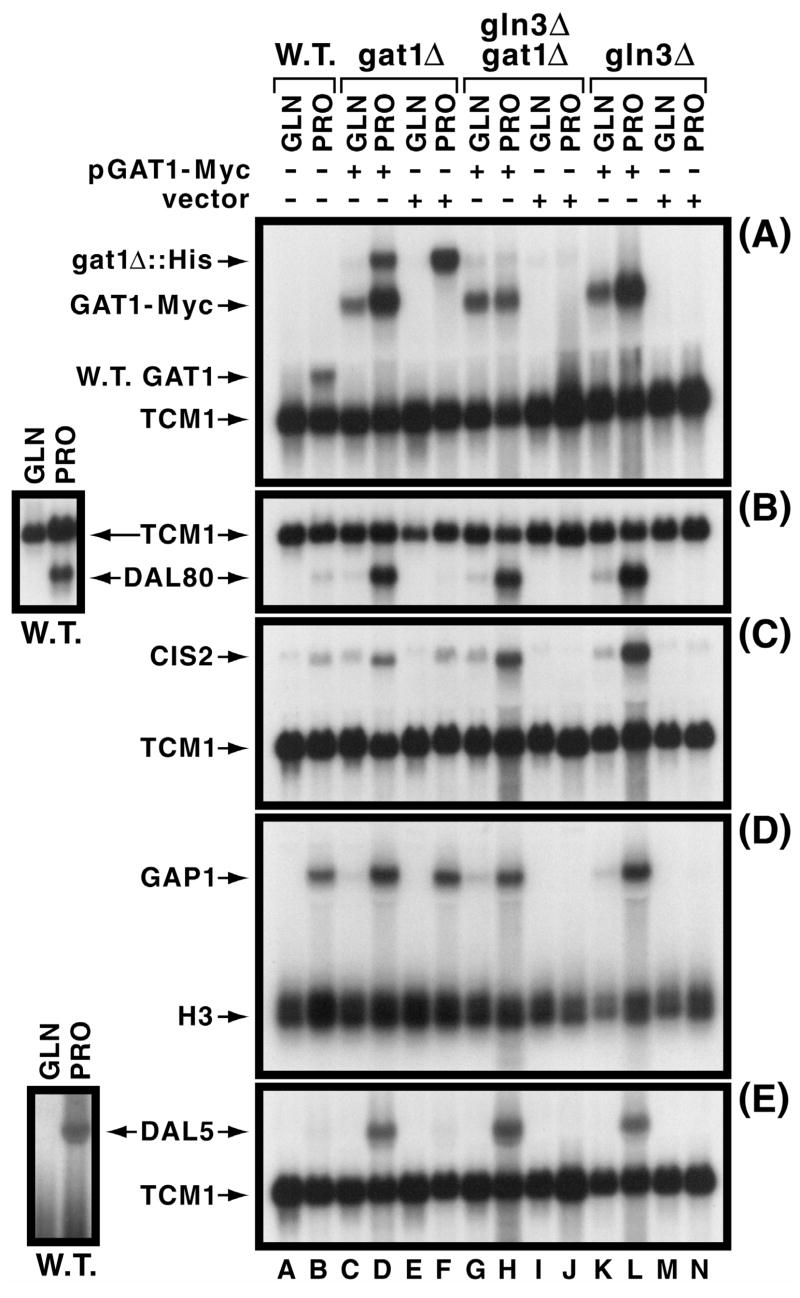
Fig. 1.
Characterization of NCR-sensitive gene expression supported by Gat1-Myc13. Panel A. GAT1 and GAT1-MYC13 expression supported by pGAT1-Myc (pKA62) or vector (pKA61) as indicated. Untransformed wild type (TCY1) and gat1Δ [(RJ715) (lanes C, D, E and F)], gln3Δ gat1Δ [(yKHC7) (lanes G, H, I and J)] or gln3Δ [(yKHC2) (lanes K, L, M and N)] transformed with pKA62 or pKA61 were grown to mid-log phase in YNB medium with either 0.1% glutamine (GLN) or proline (PRO). A GAT1-specific probe was used to detect expression of GAT1-MYC, with the TCM1 probe being used to assess uniform loading and transfer efficiencies. Panels B–E. Assessment of pGAT1-Myc’s ability to support NCR-sensitive expression of DAL80, CIS2, GAP1 or DAL5. Strains and conditions were as in Panel A. TCM1 and H3 were used as in Panel A. Small images left of the main figure depict a second experiment characterizing wild type NCR-sensitive DAL80 and DAL5 expression. This was done because proper exposure of the weak signals for these two genes would have resulted in over-exposure of the other species in the figure to the point of being impossible to critically assess NCR-sensitivity. A similarly longer exposure would likely also be required to demonstrate GAP1 expression that occurs in proline-grown gln3Δ cultures (lane N).