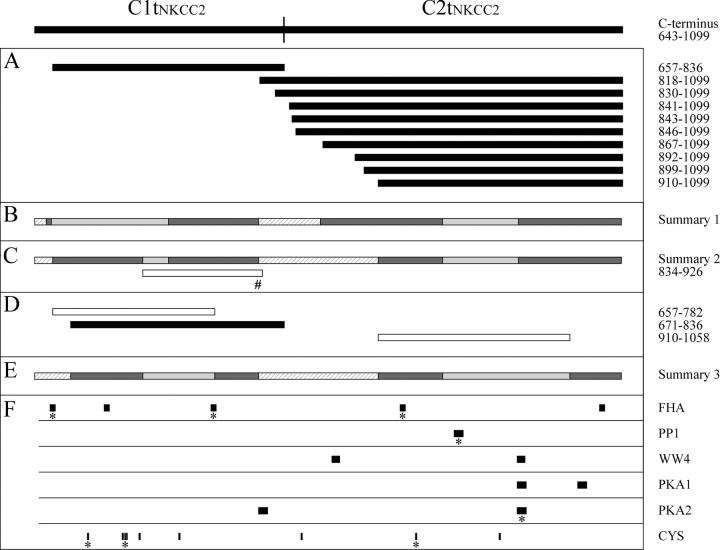
Figure 3.
huNKCC2 protein segments represented schematically according to their position in the COOH terminus and their ability to support an interaction. Each horizontal bar corresponds to a huNKCC2 residue or protein segment aligned with the region of Ct to which it corresponds. (A, B, C#, and D) Localization of Ct1 segments coexpressed in yeast with Ct2NKCC2(830–1099) or Ct2NKCC2(841–1099) and of Ct2 segments coexpressed with Ct1NKCC2(657–836). Here, black bars are used to indicate that the protein segment supports an interaction, white bars, that it does not, and the pound sign, that it was analyzed in a previous study. (C) Properties of various protein segments based on previous mapping analyses (Simard et al., 2004a) and on the double hybrid screen. Such properties are illustrated as follows: hatched bars indicate that the protein segment is not required for the Ct1–Ct2 association, light gray bars, that it may or may not be required, and dark gray bars, that it is minimally required but not sufficient; the dark gray bars are referred to in the manuscript as essential interacting regions (EIRs). (E) Properties of protein segments based on the behavior of three additional cotransformants. (F) Positions of residues or consensus binding sites in the huNKCC2 Ct. Here, the asterisks indicate that the residues or consensus sites are conserved between NKCC1 and 2, and the abbreviations FHA, PP1, WW4, PKA1, and PKA2 signify forkhead-associated binding domain, protein phosphatase type 1, [ST]P-containing binding domain, protein kinase A type, and protein kinase A type 2.