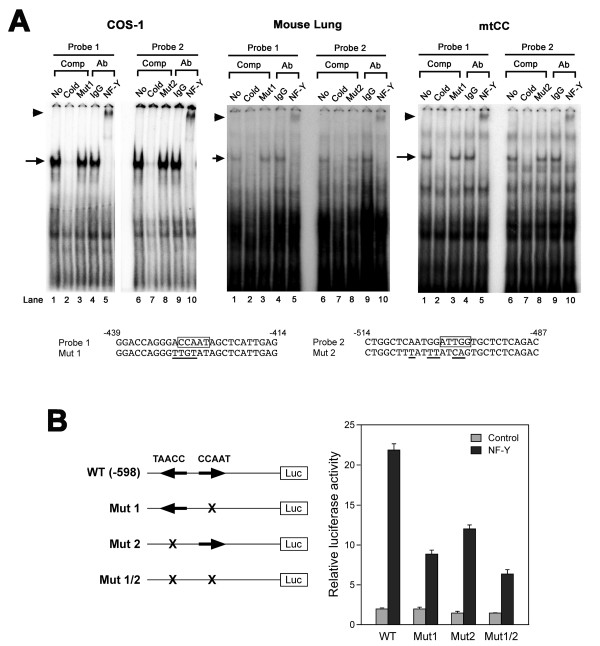
Figure 3.
Analysis of NF-Y binding sites. (A) EMSA analysis of NF-Y binding sites. Proximal (Probe 1: -425 to -429 bp, lane 1–5) and distal (Probe 2: -498 to -502 bp, lane 6–10) "CCAAT" binding sites in the mouse Scgb3a1 gene promoter were incubated with nuclear extracts prepared from COS-1 cells over-expressing NF-Y, mouse lung and mtCC cells in the presence of no competitor (lane 1, 6), 200-fold excess of own cold oligonucleotide as a competitor (lane 2, 7), 200-fold excess of mutant 1 or 2 as a competitor (lane 3 or 8, respectively), rabbit IgG as a control antibody (lane 4, 9), and polyclonal anti-NF-Y (anti-NF-YA and NF-YB combined) (lane 5, 10). NF-Y-DNA complex is indicated by an arrow, and NF-Y supershifted band is indicated by an arrowhead. Sequences for probe 1 and 2, and their mutants are shown at the bottom. These mutations were designed based on [37] and modification was added until complete abolishment of NF-Y binding was obtained. "CCAAT" sites are boxed and the mutated bases are underlined. (B) Transfection analysis of mutant constructs. COS-1 cells were co-transfected with -598 wild-type or a mutant construct (Mut 1, Mut 2) in the presence or absence of NF-Y expression plasmid. An arrow indicates the direction of the "CCAAT" sequence. Relative luciferase activity is shown as the mean ± SD based on the activity of wild-type construct -598 in the absence of NF-Y as 1 from three independent experiments, each carried out in duplicate.