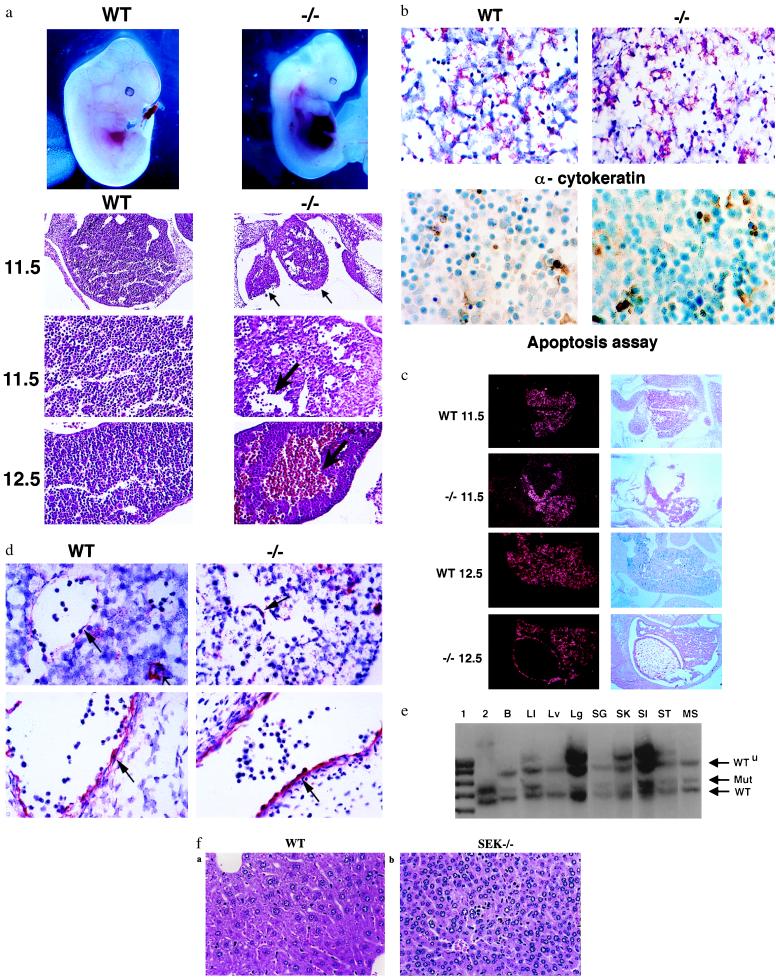
Figure 3.
Characterization of SEK1−/− embryos. (a) (Top) Wild-type (WT) and SEK1−/− (−/−) embryos at E12.5. (Bottom) Histological analysis with hematoxylin and eosin staining. Wild-type and SEK1−/− liver at E11.5 (×10 and ×25) and E12.5 (×25). (b) (Top) Immunohistochemical analysis of wild type (WT) and SEK1−/− (−/−) embryos at E11.5 with mAb to cytokeratin (Dako) (×63). The red color indicates specific staining. The numbers of cytokeratin-expressing cells per high power field are 476 ± 34, 318 ± 63, 350 ± 44, and 318 ± 17 for livers from wild-type E12.5, SEK1−/− E12.5, wild-type E11.5, and SEK1−/− E11.5 embryos, respectively. (Bottom) ApopTag staining of sections from wild-type (WT) and SEK1−/− (−/−) embryos at E12.5 (×63). The brown color indicates specific staining. The number of apoptotic cells per high power field was 34 ± 1 and 66 ± 13 for wild-type and SEK1−/− embryos, respectively. (c) In situ hybridization studies for transthyretin mRNA expression at day 11.5 and 12.5. (Right) Hematoxylin and eosin staining of corresponding sections. (d) Immunohistochemical analysis of wild-type (WT) and SEK1−/− (−/−) embryos at E11.5 with polyclonal antisera to Factor VIII (Dako) (×63). (Top) Liver. The small arrow delineates megakaryocytes. The large arrow shows the endothelial cells. (Bottom) Control section on other regions of the embryo. (e) Representative Southern blot analysis of tissues from a highly chimeric mouse. SEK1−/− cells fail to contribute to the liver. Five Chimeras were analyzed. Lanes: 1, marker; 2, phage DNA containing the SEK1 gene; B, bladder; LI, large intestine; Lv, liver; Lg, lung; SG, salivary gland; SK, skin; SI, small intestine; ST, stomach; MS, muscle. (f) Histopathology of liver derived from a highly chimeric adult mouse. Stained with hematoxylin and eosin. (a) Normal liver (×40). (b) SEK1−/− chimera (×40). Note the nuclear fragmentation and abnormal hepatocyte lobule structure.