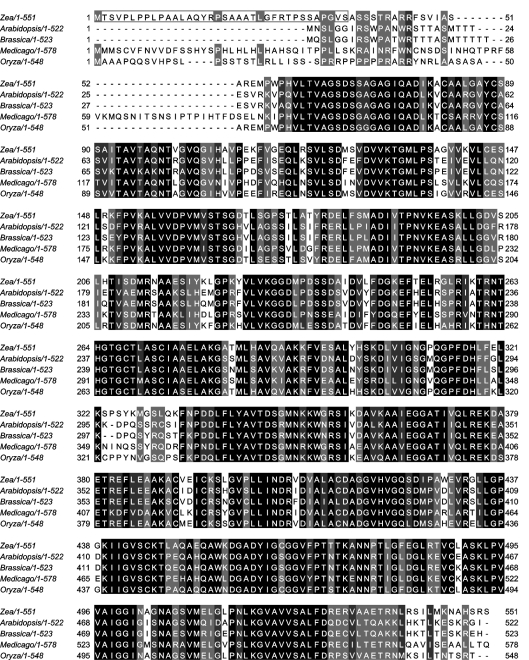
Figure 4. Multiple alignment of the amino acid sequences of THI3 and its orthologues occurring in various plant species.
The accession codes of the sequences compared were emb/CAJ45026 (Z. mays, the present study), gb/AAM91567 (A. thaliana), gb/AAC31298 (B. napus, BTH1), gb/ABE87727 (M. truncatula) and gb/ABA96049 (O. sativa). The alignment was performed using the ClustalW program and the result visualized in JalView. Black, dark grey and light grey highlights mark the positions in the primary structures where all 5, 4 out of 5 or 3 out of 5 residues are identical respectively. The box at the N-terminus of Z. mays THI3 protein includes the putative 36-amino-acid residue chloroplast transit peptide predicted by the ChloroP program [25].