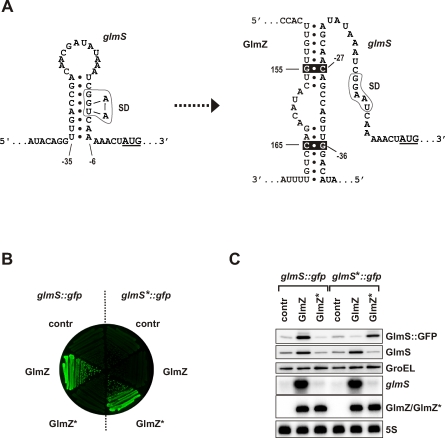
Figure 3. GlmZ Directly Acts on the glmS mRNA to Enhance GlmS Synthesis.
(A) Predicted anti-antisense mechanism by which GlmZ activates the E. coli glmS mRNA. The left panel shows the 5′ UTR of the glmS mRNA in an unbound, “inactive” state in which an intrinsic hairpin sequesters the glmS SD (boxed) and thus inhibits translation. Residues −42 to +3 relative to the AUG start codon, which is underlined, are shown. The right panel shows glmS mRNA in its “activated” form, i.e., upon base-pairing of residues 150–157 and 163–169 of GlmZ RNA with the glmS 5′ UTR, which disrupts the inhibitory hairpin and liberates the glmS SD. The positions of point mutations introduced in glmS (C−27 → G, G−36 → C; glmS* allele) and GlmZ (G+155 → C, C+165 → G; GlmZ* allele), which are expected to maintain base pairing in the glmS*/GlmZ* duplex, are indicated by inverse (white on black) print.
(B) Colony fluorescence of strain JVS-8113 (ΔglmY ΔglmZ) carrying either the glmS::gfp or glmS*::gfp fusion plasmids, each in combination with plasmid pJV300 (“contr”), pPL-GlmZ (“GlmZ”), or pPL-GlmZ* (“GlmZ*”). Shown is an image of a LB agar plate obtained in the fluorescence mode.
(C) The strains shown in (B) were grown to early stationary phase and subjected to western blot (top three panels) and northern blot (bottom three panels) analysis. The plasmid-encoded GlmS::GFP fusion protein was detected using α-GFP antibodies (top panel); the chromosomally encoded glmS mRNA and GlmS protein, and the plasmid-encoded GlmZ RNA (as indicated) were detected as in Figure 2.