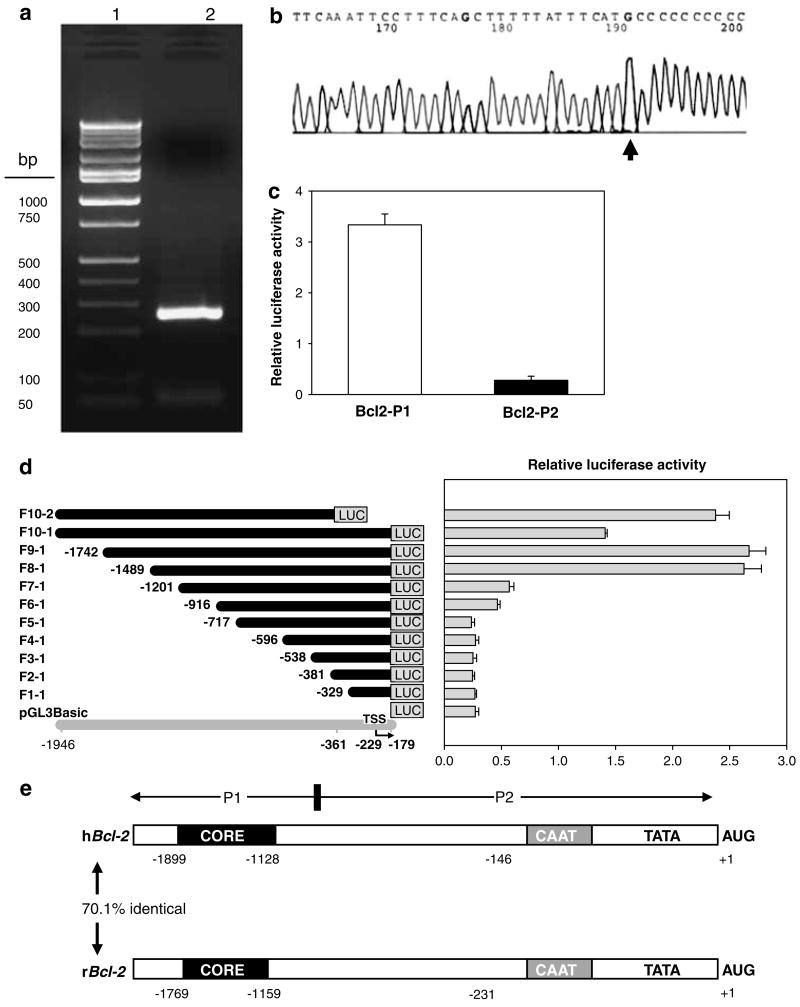
Figure 1.
Characterization of the rat Bcl-2 promoter. (a) Product from 5′-rapid amplification of cDNA ends (5′-RACE) on a 2% agarose gel stained with ethidium bromide. In 5′-RACE, fresh poly(A)+RNA (Ambion) was used to ensure a high percentage of full-length transcripts, which consistently generated a single 260 bp PCR product (lane 2; lane 1=marker ladder). Degradation of full-length poly(A)+RNAs was discounted based on the lack of shorter or longer 5′-RACE products in repeat experiments, and DNA sequencing (GenBank accession no. AF531426) indicated a common 5′ end with the major TSS assigned to a C nucleotide 231 bp upstream of the translation start site (b), consistent with the human BCL-2 counterpart. (c) Bcl-2 promoter-luciferase reporter construct containing promoter region P1 had >10-fold higher activity than region P2 when transfected into HEK293 cells and luciferase activities were measured in whole cell lysates 48 h post-transfection. (d) Deletion analysis. Left panel: schematic view of Bcl-2 promoter-luciferase reporter constructs; gray line, position in genomic DNA sequence; bent arrow, transcription start site (TSS); black lines, extent of genomic DNA attached to the luciferase (LUC) gene. Promoter fragments were designated as F1-1 through F10-2, as shown at left. Right panel: Bcl-2 promoter activity in HEK293 cells. Transcriptional activity was evaluated by transient transfection, using the constructs shown in the left panel. Cells were extracted using 1×Reporter lysis buffer, 48 h post-transfection. Values represent the mean±s.d. of three independent transfections for each construct, normalized for transfection efficiency using pSV-β-Gal, as reported before (Li et al., 2004). (e) Genomic organization of human BCL-2 (hBCL-2) and rat Bcl-2 (rBcl-2) promoters, with numbering relative to the translation start codon (AUG). CORE, core promoter region.