Fig. 6.
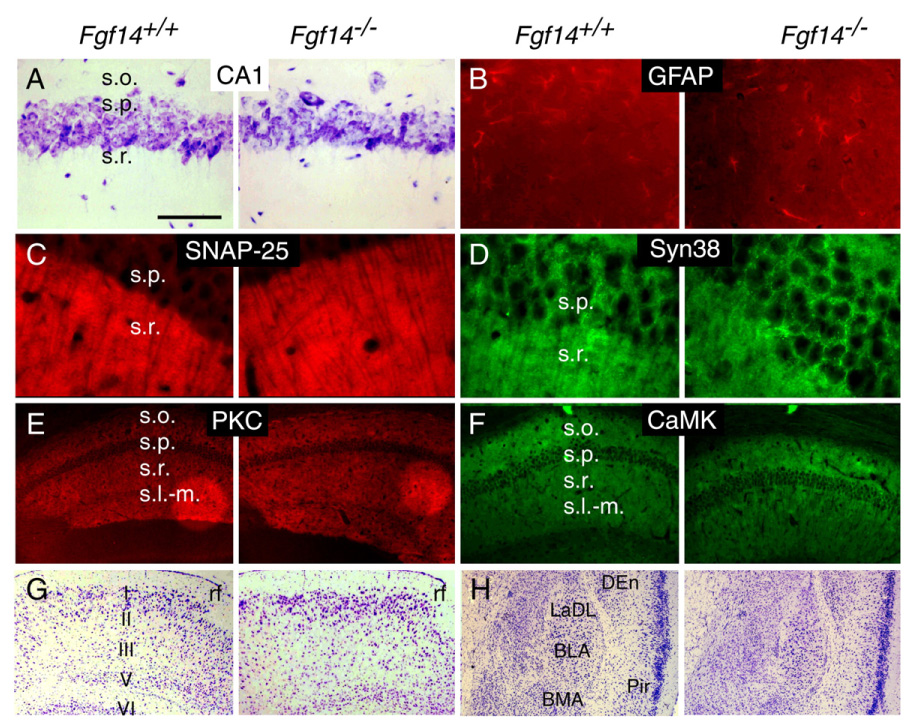
Normal PHR and amygdala anatomy in Fgf14−/− mice. (A) Normal hippocampal cytoarchitecture in the s.o., s.p. and s.r. of CA1 revealed by Nissl staining in Fgf14−/− and WT mice. (B–F) No difference was observed in the staining intensity of GFAP (B), SNAP-25 (C), Synaptophysin (Syn38) (D), PKCγ (E) and CaMKII (F) in CA1 regions of Fgf14−/− and WT mice. (G–H) Normal cytoarchitecture in the entorhinal cortex (G) and amygdala (H) in WT and Fgf14−/− mice (Nissl stain). BLA, anterior basolateral amygdaloid nucleus; BMA, anterior basolateral amygdala; LaDL, dorsolateral amygdaloid nucleus DEn, dorsal endopiriform nucleus; Pir, piriform cortex; rf, rhinal fissure; s.l-m., stratum lacunosum moleculare; s.o., stratum oriens; s.p., stratum pyramidal; s.r., stratum radiatum. Bar=125 µm (A, B); Bar=50 µm (C, D); Bar=500 µm (E–H).
