Figure 3.
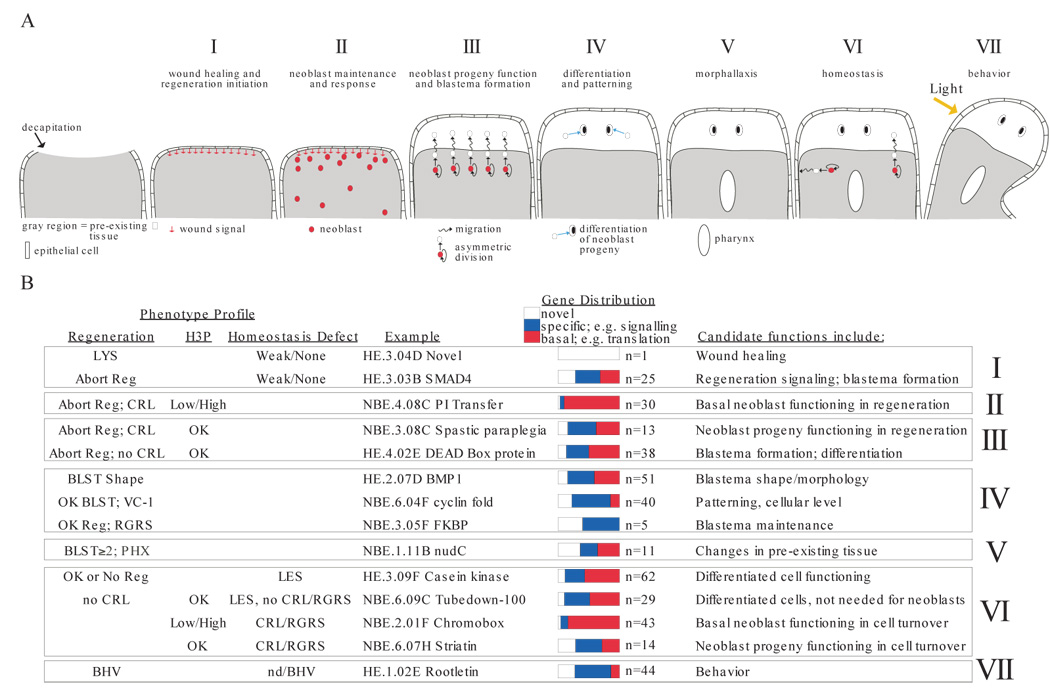
Identification of genes with candidate functions in planarian regeneration. (A) Planarian regeneration is divided into seven stages, “I” through “VII”. Anterior region of a decapitated planarian is shown, facing up. Wound healing involves epidermal cell spreading. The blastema is anterior and white. Stages correspond to numbered text sections in which phenotype categories are discussed. Neoblasts are depicted only when a process involving their function is described. V, pharynx formation occurs in pre-existing tissue of a fragment lacking original pharynx. (B) Gene groups sharing phenotype profiles are summarized. Profiles identify genes with predicted functions corresponding to regeneration stages in (A). Some genes are found in multiple categories. LYS, lysis. Reg, regeneration (blastema formation); “abort”, too small or no blastema. CRL, curling. BLST, blastema. VC-1, abnormal photoreceptors (see text, Table S3). PHX, pharynx regeneration in tail fragments. RGRS, tissue regression. BHV, behavior abnormal. H3P categorization as in Figure 4, Table S3. LES; lesions. Novel, no predicted function. Specific, if predicted to encode proteins involved in signal transduction, transcription, cell adhesion, neuronal functions, disease, RNA binding, channels/transporter function, cytoskeletal regulation. Basal, if predicted to encode proteins involved in translation, metabolism, RNA splicing, proteolysis, protein folding, vesicle trafficking, cell cycle, or cytoskeleton machinery.
