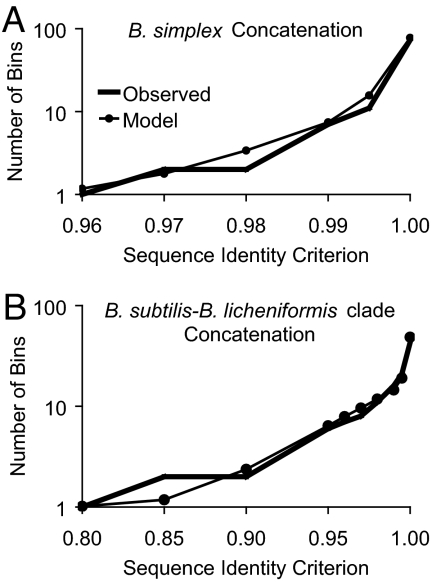
Fig. 1.
Observed and modeled clade sequence diversity patterns. Sequences for a gene (or a concatenation of genes) were binned by complete linkage clustering (23) into clusters with different levels of minimum pairwise identity. ES analysis of each clade yielded two solutions, one with low drift and one with high drift. The high-drift solutions require population sizes that are unrealistically low for these taxa (SI), so the model curves are based on the low-drift solutions. For taxa with low population sizes and/or extremely high recombination rates, high-drift solutions (with little or no periodic selection) may be the most appropriate. The individual points for each model are means based on 1,000 replications of the low-drift solution. (A) Diversity among 116 B. simplex isolates from Evolution Canyons I and II based on a concatenation of gapA, rpoB, and uvrA, with recombinant organisms removed. (B) Diversity among 73 isolates within the B. licheniformis–B. subtilis clade, primarily from Evolution Canyon III, as based on a concatenation of gapA, gyrA, and rpoB, with recombinant organisms removed.