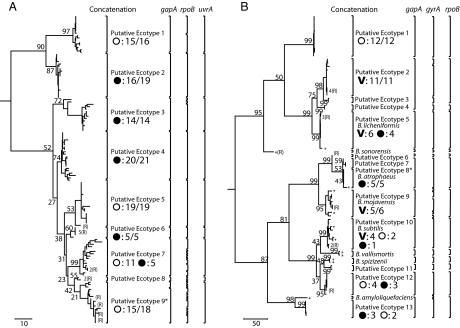
Fig. 3.
Phylogeny and ecotype demarcation of the B. simplex and B. subtilis–B. licheniformis clades. Phylogenies are based on parsimony, with 100-replicate bootstrapping, using MEGA (37). In isolates for which one gene had recombined (a group of related recombinants is indicated by “R” following the number of recombinants), the recombined gene was replaced in the input for MEGA by “unknown” nucleotides; so the phylogeny estimates the recombination-free tree. Ecotypes were demarcated conservatively as the most inclusive clades that were each consistent with being a single ecotype (SI). For nearly all putative ecotypes, the maximum likelihood solution of n was equal to 1, although in some cases n = 2 with the lower confidence interval including n = 1. Ecotype demarcations are indicated by brackets, for the concatenation and each individual gene. The ecotype demarcations were similar based on the concatenation and the individual genes, except that the more rapidly evolving genes (gyrA in the case of B. subtilis–B. licheniformis and rpoB in the case of those genes analyzed in B. simplex) tended to split the ecotypes determined by analysis of the concatenation. For isolates that had recombined at one gene locus, ecotype placement was determined by ES of the two genes that had not recombined. For example, the three recombinants indicated within PE 5 of the B. subtilis–B. licheniformis clade (which had recombined at gapA) were found to be in the same ecotype as the members of PE 5 in analyses of gyrA and rpoB. With two exceptions, demarcated ecotypes were supported as monophyletic groups in at least 50% of bootstrap replications; the exceptions were PE9 of B. simplex and PE8 of the B. subtilis–B. licheniformis clade, which are asterisked to indicate that their phylogenetic status is tentative, pending additional sequence data. The median bootstrap support for putative ecotypes was 72% within B. simplex and 99% within the B. subtilis–B. licheniformis clade. Whereas the B. simplex ecotypes do not generally have high bootstrap support, owing in part to their very close relatedness, their monophyly is supported by alternative phylogenetic approaches, including Bayesian and neighbor joining algorithms (data not shown). Microhabitat sources were the south-facing slope (open circles), the north-facing slope (filled circles), and the canyon bottom (indicated by V). For each ecotype represented by at least four isolates, the principal microhabitat source(s) is indicated. If one microhabitat provided at least 80% of the isolates, the principal microhabitat source is indicated; for ecotypes not so dominated by a single source, all microhabitat sources are indicated. The number of isolates from each source is indicated. (A) The phylogeny of B. simplex rooted by B. subtilis, B. licheniformis, B. cereus, and B. halodurans. The nine putative ecotypes of this clade differ significantly in their associations with the two slopes. (B) The phylogeny of the B. subtilis–B. licheniformis clade rooted by B. halodurans. The 13 putative ecotypes differ significantly in their associations with the two slopes and the canyon bottom.