Abstract
In the Drosophila embryo, body wall muscles are formed by the fusion of two cell types, Founder Cells (FCs) and Fusion Competent Myoblasts (FCMs). Using an enhancer derived from the Dmef2 gene ([C/D]*), we report the first GAL4 driver specifically expressed in FCMs. We have determined that this GAL4 driver causes expression in a subset of FCMs and, upon fusion, in developing myotubes from stage 14 onwards. In addition, we have shown that using this Dmef2-5x[C/D]*-GAL4 driver to express dominant negative Rac in only FCMs causes a partial fusion block. This novel GAL4 driver will provide a useful reagent to study Drosophila myoblast fusion and muscle differentiation.
Keywords: Muscle, Fusion Competent Myoblast, Myotube, Drosophila, Dmef2, Lmd, Rac, Myoblast, Fusion
1. Results and Discussion
Dmef2 is the sole Drosophila member of the Myocyte Enhancing Factor 2 (MEF2) family of transcription factors (Nguyen et al., 1994). Dmef2 is required for the differentiation of mesodermal tissues within the developing Drosophila embryo, specifically the body wall, visceral and cardiac muscles (Bour et al., 1995; Lilly et al., 1995; Lin et al., 1996; Ranganayakulu et al., 1995). During the early stages of mesoderm development, Dmef2 is expressed in all mesodermal cells and its expression is controlled by another mesodermal transcription factor, Twist (Twi) (Cripps et al., 1998). Dmef2 expression continues in somatic, visceral and cardiac progenitors and is maintained throughout the development of these tissues (Bour et al., 1995; Lilly et al., 1994; Ranganayakulu et al., 1995). Previous studies identified enhancers responsible for the expression of Dmef2 in the two cell types that form body wall muscles, the Founder Cells (FCs) and Fusion Competent Myoblasts (FCMs) (Nguyen and Xu, 1998). FCs determine the identity of individual muscles. Upon fusion to FCs, FCMs become entrained to the FCs individual developmental program (Baylies et al., 1998; Beckett and Baylies, 2006a). Further dissection of an FCM-specific enhancer (I-E) identified a 55bp sequence ([C/D]*) that is necessary and sufficient to direct expression of a lacZ reporter gene in FCMs. This [C/D]* enhancer is bound directly by the FCM-specific transcription factor Lame duck (Lmd, also known as Myoblasts incompetent and Gleeful), requires lmd for its expression and therefore mediates FCM-specific expression of Dmef2 (Duan et al., 2001; Furlong et al., 2001; Ruiz-Gomez et al., 2002) . We have cloned multiple copies of this [C/D]* enhancer element upstream of the GAL4 gene to create a novel GAL4 driver (Dmef2-5x[C/D]*-GAL4) that we characterize here.
We crossed flies homozygous for the Dmef2-5x[C/D]*-GAL4 driver to flies carrying a UAS-GFP transgene and analyzed GFP expression in the resulting embryos. We detected mesoderm-specific GFP expression from stage 14 onwards (10.5 hrs after egg laying [AEL]; Fig. 1). GFP expression is first detected during early to mid stage 14 in a small number of ventral mesodermal cells (Fig. 1A). This initiation of reporter gene expression is later than that observed using a direct lacZ fusion (Duan et al., 2001) and presumably is due to the delay caused by the GAL4/UAS system. From late stage 14 and throughout stage 15 (11.5-13 hrs AEL), GFP expression is detected along the dorsal-ventral (D-V) axis of the embryo in both growing myotubes and surrounding cells (Fig. 1B). Based on their location, it is likely that these surrounding cells are FCMs. GFP expression is also detected in the final body wall muscles at stage 16 (13-16 hrs AEL; Fig. 1C-D). However, when we analyzed these embryos we found that GFP expression was not detected in all muscles and GFP levels varied between muscles. This differential muscle expression is variable between hemisegments and embryos, and is quantified in Table 1. For example, Fig. 1D shows that GFP is not expressed in all LT muscles, and in those in which it is expressed, it is not expressed at the same level. We also found that GFP expression is rarely detected in the VT1 and VA3 muscles. These muscles are both small in size, with the VT1 muscle, for example, containing 2-4 nuclei at stage 16 (Bate, 1990; Beckett and Baylies, 2007). The absence of GFP expression could be due to the late initiation of GFP expression in FCMs as well as the small number of fusion events occurring in these muscles. The expression of GFP in mesodermal cells surrounding the growing myotubes and the varying levels of GFP expression in the final muscles suggested that this the Dmef2-5x[C/D]*-GAL4 driver is expressed in FCMs and that, upon fusion to myotubes, is also expressed in these cells. The variation in GFP levels in the final muscles between hemisegments and embryos also suggested that the Dmef2-5x[C/D]*-GAL4 driver is not expressed in all FCMs, consistent with the temporal delay caused by the GAL4/UAS system, and that an individual FCM does not always fuse to the same muscle.
Figure 1. The Dmef2-5x[C/D]*-GAL4 driver causes expression of a GFP reporter in the somatic mesoderm and body wall muscles.
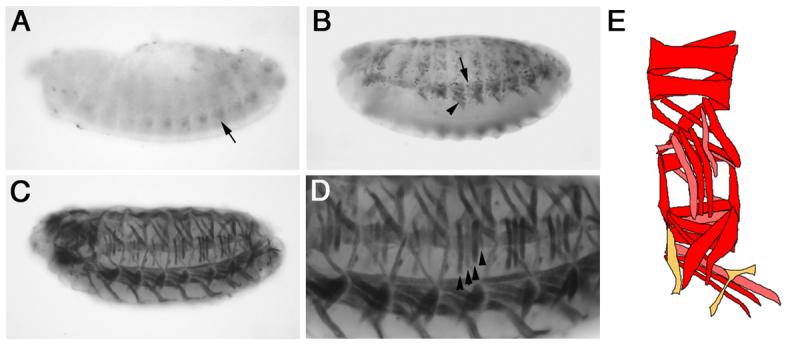
Dmef2-5x[C/D]*-GAL4 > UAS-GFP embryos were stained with an antibody against GFP. Lateral views of stage 14 (A), 15 (B) and 16 (C-D) embryos are shown. Panel D shows a close up of the embryo in panel C. (A) In contrast to the direct enhancer-reporter construct (Duan et al, 2001), GFP expression is first detected in a subset of cells in the ventral somatic mesoderm at during stage 14 (black arrow, A). (B) During stage 15, GFP expression is detected in growing myotubes (black arrowhead, B) and surrounding cells (black arrow, B). (C-D). GFP expression is detected in the final body wall muscles at stage 16. GFP is not detected in all muscles or at uniform levels. Black arrowheads highlight differing levels of GFP expression in LTs1-4. (E) Schematic showing quantification of GFP expression in individual muscles from Table 1. Red = 75-100%, pink = 50-74% and orange = <50%.
Table 1.
Expression of Dmef2-5x[C/D]*-GAL4 > UAS-GFP in final larval body wall muscles.
| Muscle | % GFP positive (n) | Muscle | % GFP positive (n) | Muscle | % GFP positive (n) |
|---|---|---|---|---|---|
| DA1 | 97% (36) | LT1 | 74% (46) | VL1 | 87% (52) |
| DA2 | 83% (36) | LT2 | 76% (49) | VL2 | 58% (53) |
| DA3 | 88% (43) | LT3 | 76% (49) | VL3 | 62% (52) |
| DO1 | 100% (36) | LT4 | 53% (45) | VL4 | 62% (53) |
| DO2 | 100% (41) | LO1 | 73% (45) | VO1 | 95% (57) |
| DO3 | 85% (53) | SBM | 82% (45) | VO2 | 95% (57) |
| DO4 | 98% (53) | VA1 | 87% (55) | VO3 | 93% (57) |
| DO5 | 50% (52) | VA2 | 76% (55) | VO4 | 61% (49) |
| DT1 | 77% (53) | VA3 | 11% (53) | VO5 | 100% (47) |
| LL1 | 91% (47) | VT1 | 8% (55) | VO6 | 83% (46) |
To confirm that the Dmef2-5x[C/D]*-GAL4 driver is expressed in FCMs and, subsequently, upon fusion, in myotubes, we used confocal microscopy. We crossed Dmef2-5x[C/D]*-GAL4 flies to flies carrying a UAS-lacZ transgene and stained the resulting embryos with antibodies against β-galactosidase (βgal) and Krüppel (Kr), an FC identity transcriptional regulator (Fig. 2A; Ruiz-Gomez et al., 1997). We detected ßgal expression in a subset of mesodermal cells from stage 14, similar to the results that we obtained using the GFP reporter described above (Fig. 2A). Kr is expressed in the nuclei of a subset of FCs and developing myotubes (Ruiz-Gomez et al., 1997). We detected Kr expression in the developing LT2 and LT4 muscles at stage 14 (Fig. 2A'). In each myotube, while both nuclei are Kr-positive, only one nucleus also expresses ßgal (Fig. 2A"). ßgal is never detected in Kr-expressing FCs prior to fusion (data not shown), therefore the co-expression observed at stage 14 must be due to fusion. In agreement with this, we detected ßgal expression in a small number of surrounding cells that are presumably FCMs.
Figure 2. The Dmef2-5x[C/D]*-GAL4 driver causes expression in subsets of FCMs and growing myotubes.
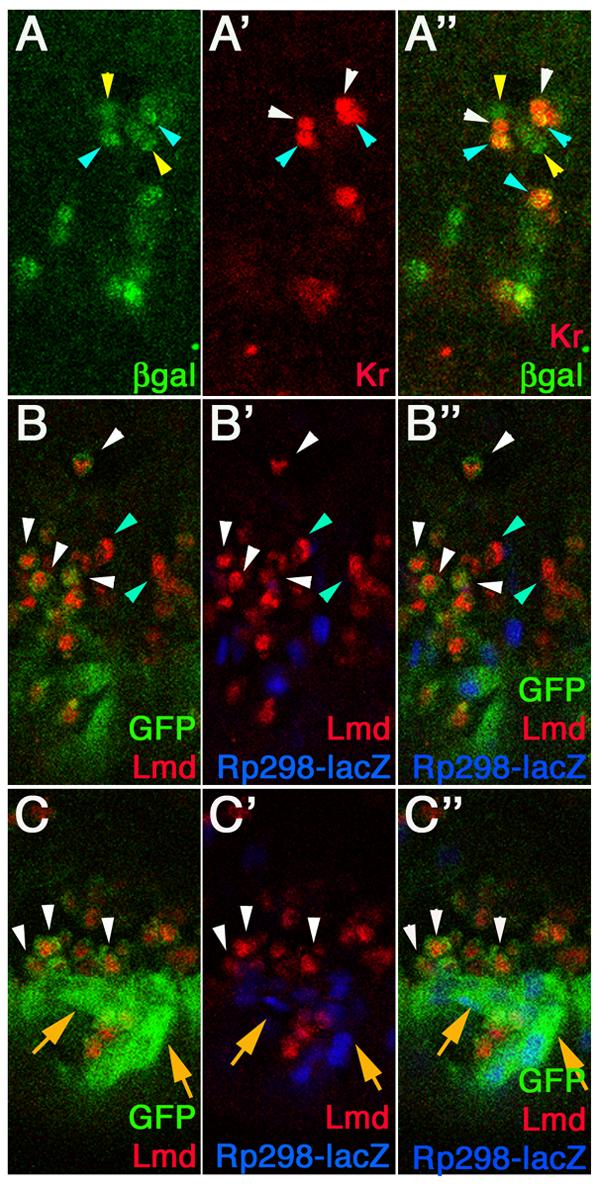
Dmef2-5x[C/D]*-GAL4 > UAS-lacZ (A) and rp298-lacZ; Dmef2-5x[C/D]*-GAL4 > UAS-2xeGFP (B-C) embryos were stained with antibodies against ßgal (green, A; blue, B-C), Kr (red, A), GFP (green, B-C) and Lmd (red, B-C). Panels A-A" show vental-lateral views of a stage 14 embryo and panels B-C show dorsal (B) and ventral (C) views of a late stage 14 embryo. (A) Kr is expressed in myotubes for the LT2 and LT4 muscles (white and blue arrowheads). ßgal is expressed in only one of the Kr-expressing nuclei in each myotube (blue arrowheads) and in surrounding Kr-negative cells (yellow arrowheads). (B) GFP is detected in a subset of Lmd-positive FCMs (white arrowheads). Not all FCMs express GFP (blue arrowheads). (C) GFP is detected in rp298-lacZ expressing myotubes (orange arrows) and surrounding FCMs (white arrowheads). Lmd expression is lost from FCMs after fusion.
To confirm that the ßgal-expressing cells not expressing FC markers are indeed FCMs, we performed colocalization experiments using an antibody against the FCM-specific protein Lmd (Duan et al., 2001). For this experiment, we crossed flies containing both the Dmef2-5x[C/D]*-GAL4 driver and the rp298-lacZ transgene, which is expressed in the nuclei of FCs and myotubes (Nose et al., 1998), to flies carrying a UAS-2xeGFP transgene. We then stained the resulting embryos with antibodies against Lmd, GFP and ßgal and analyzed them using confocal microscopy (Fig. 2B). In these embryos, we detected GFP expression in a subset of Lmd-expressing FCMs at fixed time points (Fig. 2B-C). We quantified this expression and determined that approximately 47% of Lmd-positive cells (n=229) in each hemisegment at a single point in time co-express GFP, indicating that they express the Dmef2-5x[C/D]*-GAL4 driver. We observed Lmd-expressing FCMs surrounding rp298-lacZ expressing myotubes as previously described (Beckett and Baylies, 2007; Duan et al., 2001), a subset of which co-expressed GFP (Fig. 2C). GFP was also detected in the rp298-lacZ expressing myotubes (Fig. 2C). These data confirmed that the Dmef2-5x[C/D]*-GAL4 driver is expressed in a subset of FCMs, at the time points examined, and continues to be expressed upon fusion with the developing myotubes during the final stages of muscle development.
We have shown that the Dmef2-5x[C/D]*-GAL4 driver leads to reporter expression in subsets of FCMs and myotubes from stage 14 until the end of muscle development. To determine if this GAL4 driver could be used to analyze events that occur during these stages, we tested whether overexpression of dominant negative Rac (dnRac) in FCMs would affect muscle development. Rac activity is required for myoblast fusion as loss of Rac function (Rac1,Rac2, Mtl triple mutants) lead to a myoblast fusion block (Hakeda-Suzuki et al., 2002; Richardson et al., in revision). Overexpression of dnRac throughout the mesoderm in both FCs and FCMs phenocopies the loss of function phenotype, resulting in a fusion block (Luo et al., 1994). We therefore asked whether expression of the dnRac construct only in FCMs would lead to a fusion block. We crossed flies carrying the Dmef2-5x[C/D]*-GAL4 driver to flies carrying a UAS-dnRac transgene and assayed the final muscle pattern of the resulting embryos using an antibody against Myosin heavy chain (Mhc). In these embryos, we observed a small number of unfused myoblasts indicating a partial fusion block (Fig. 3B). To confirm that this lack of fusion was due to expression of dnRac using the Dmef2-5x[C/D]*-GAL4 driver, we repeated the experiment using flies carrying both UAS-dnRac and UAS-GFP transgenes. We stained the resulting embryos with antibodies against Mhc and GFP and analyzed them using confocal microscopy. We observed the same partial fusion block and confirmed that the majority of unfused myoblasts are GFP-positive (Fig. 3C). We also observed reduced levels of GFP in the muscles of these embryos, indicating that the GFP expression observed in the final muscles of Dmef2-5x[C/D]*-GAL4 > UAS-GFP embryos shown in Figure 1 is truly due to the fusion of GFP-expressing FCMs with developing myotubes.
Figure 3. Expression of dominant negative Rac using Dmef2-5x[C/D]*-GAL4 causes a partial fusion block.
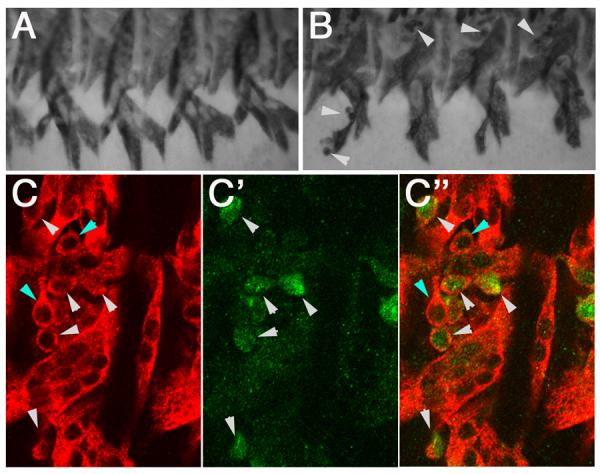
Stage 16 wild type (A), Dmef2-5x[C/D]*-GAL4 > UAS-dnRac (B) and Dmef2-5x[C/D]*-GAL4 > UAS-dnRac; UAS-GFP (C) embryos stained with antibodies against Mhc (A-B, red in C) and GFP (green in C) are shown. (A) Wild type embryos show a segmentally repeated pattern of ventral muscles. No unfused myoblasts are observed at this stage. (B) Small numbers of unfused myoblasts are observed in Dmef2-5x[C/D]*-GAL4 > UASdnRac embryos (white arrowheads). (C) The majority of unfused myoblasts in Dmef2-5x[C/D]*-GAL4 > UAS-dnRac; UAS-GFP embryos express GFP (white arrowheads). A small number of non-GFP unfused myoblasts (blue arrowheads) are also observed. Low levels, if any, of GFP expression is detected in the muscles of these embryos.
To conclude, we have constructed and characterized a novel FCM-GAL4 driver that is expressed specifically in a subset of FCMs at the time points examined and, post-fusion, in the majority of developing myotubes from stage 14 until the end of muscle development. We have further shown that this tool can be used to block fusion and in a parallel study to block muscle attachment (Beckett and Baylies, submitted). To our knowledge, this is the first FCM-specific GAL4 driver to be described and will provide an extremely useful tool for the study of fusion and late events during muscle differentiation.
2. Experimental Procedures
2.1. Drosophila genetics
Fly strains used were: Dmef2-5x[C/D]*-GAL4 (this study), UAS-GFP (Bloomington), UAS-2xeGFP (a gift from A. Michelson), UAS-lacZ (Bloomington), rp298-lacZ (Nose et al., 1998), UAS-RacL89.6 (Luo et al., 1994). All experiments were performed at 25°C using standard genetic crosses.
2.2. Construction of Dmef2-5x[C/D]*-GAL4 flies
A 55bp sequence ([C/D*]) located in the upstream region of Dmef2 was previously identified as a minimal enhancer sufficient for FCM expression (Duan et al., 2001). 5 copies of the [C/D*] fragment were ligated with uniform directionality from annealed oligos with AvaI sites and then cloned into the NotI site of the pDA188 vector (kindly provided by Konrad Basler). The constructed plasmid was used to generate Dmef2-5x[C/D*]-GAL4 transgenic flies using established protocols (Nguyen and Xu, 1998).
2.3. Immunohistochemistry
All embryos were fixed using 4% paraformaldhyde/heptane. Antibodies were preabsorbed 1:10 against fixed wild type embryos (PA) or in combination with the TSA system (TSA; PerkinElmer Life Sciences) where stated. For DAB staining, the antibody dilutions used were: mouse anti-GFP (1:200; PA; Clonetech) and rabbit anti-Mhc (1:20,000; TSA; a gift from D. Keihart). Biotinylated secondary antibodies were used in combination with Vector Elite ABC kit (Vector Laboratories, CA). Specimens were embedded in Araldite and images captured using an Axiocam camera (Zeiss) and processed using Adobe Photoshop 7.0. For fluorescent staining, the antibody dilutions used were: mouse anti-ßgal (1:2000; Promega), chicken anti-ßgal (1:1000; Abcam), guinea pig anti-Kr (1:500; PA; a gift from J. Reinitz), mouse anti-GFP (1:200; PA; Clonetech), rabbit anti-Lmd (1:250; PA) and rabbit anti-Mhc (1:1000; a gift from D. Kiehart). Alexa488, Alexa555 and Alexa647 conjugated secondary antibodies were used (1:400; Molecular Probes) and embryos were mounted in Vectashield (Vector Laboratories, CA). Embryos were analyzed using a Zeiss LSM 510 confocal scanning system. Images were exported from Zeiss LSM software and processed using Adobe Photoshop 7.0.
3. Acknowledgements
We wish to thank J. Reinitz, A. Michelson, K. Basler and D. Kiehart for reagents. This work was supported by the Sloan Kettering Institute and NIH grants (GM586989, GM78318 to M.K.B and AR4628 to H.T.N).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bate M. The embryonic development of larval muscles in Drosophila. Development. 1990;110:791–804. doi: 10.1242/dev.110.3.791. [DOI] [PubMed] [Google Scholar]
- Baylies MK, Bate M, Ruiz Gomez M. Myogenesis: a view from Drosophila. Cell. 1998;93:921–927. doi: 10.1016/s0092-8674(00)81198-8. [DOI] [PubMed] [Google Scholar]
- Beckett K, Baylies MK. The development of the Drosophila larval body wall muscles. Int Rev Neurobiol. 2006a;75:55–70. doi: 10.1016/S0074-7742(06)75003-6. [DOI] [PubMed] [Google Scholar]
- Beckett K, Baylies MK. Parcas, a regulator of non-receptor tyrosine kinase signaling, acts during anterior-posterior patterning and somatic muscle development in Drosophila melanogaster. Dev Biol. 2006b;299:176–192. doi: 10.1016/j.ydbio.2006.07.049. [DOI] [PubMed] [Google Scholar]
- Beckett K, Baylies MK. 3D analysis of founder cell and fusion competent myoblast arrangements outlines a new model of myoblast fusion. Dev Biol. 2007 doi: 10.1016/j.ydbio.2007.06.024. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckett K, Baylies MK. Multiple roles for Src during Drosophila myogenesis: Links to Twist, EGFR and Integrin signaling. submitted. [Google Scholar]
- Bour BA, O'Brien MA, Lockwood WL, Goldstein ES, Bodmer R, Taghert PH, Abmayr SM, Nguyen HT. Drosophila MEF2, a transcription factor that is essential for myogenesis. Genes Dev. 1995;9:730–741. doi: 10.1101/gad.9.6.730. [DOI] [PubMed] [Google Scholar]
- Cripps RM, Black BL, Zhao B, Lien CL, Schulz RA, Olson EN. The myogenic regulatory gene Mef2 is a direct target for transcriptional activation by Twist during Drosophila myogenesis. Genes Dev. 1998;12:422–434. doi: 10.1101/gad.12.3.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan H, Skeath JB, Nguyen HT. Drosophila Lame duck, a novel member of the Gli superfamily, acts as a key regulator of myogenesis by controlling fusion-competent myoblast development. Development. 2001;128:4489–4500. doi: 10.1242/dev.128.22.4489. [DOI] [PubMed] [Google Scholar]
- Furlong EE, Andersen EC, Null B, White KP, Scott MP. Patterns of gene expression during Drosophila mesoderm development. Science. 2001;293:1629–1633. doi: 10.1126/science.1062660. [DOI] [PubMed] [Google Scholar]
- Hakeda-Suzuki S, Ng J, Tzu J, Dietzl G, Sun Y, Harms M, Nardine T, Luo L, Dickson BJ. Rac function and regulation during Drosophila development. Nature. 2002;416:438–442. doi: 10.1038/416438a. [DOI] [PubMed] [Google Scholar]
- Lilly B, Galewsky S, Firulli AB, Schulz RA, Olson EN. D-MEF2: a MADS box transcription factor expressed in differentiating mesoderm and muscle cell lineages during Drosophila embryogenesis. Proc Natl Acad Sci U S A. 1994;91:5662–5666. doi: 10.1073/pnas.91.12.5662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilly B, Zhao B, Ranganayakulu G, Paterson BM, Schulz RA, Olson EN. Requirement of MADS domain transcription factor D-MEF2 for muscle formation in Drosophila. Science. 1995;267:688–693. doi: 10.1126/science.7839146. [DOI] [PubMed] [Google Scholar]
- Lin MH, Nguyen HT, Dybala C, Storti RV. Myocyte-specific enhancer factor 2 acts cooperatively with a muscle activator region to regulate Drosophila tropomyosin gene muscle expression. Proc Natl Acad Sci U S A. 1996;93:4623–4628. doi: 10.1073/pnas.93.10.4623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L, Liao YJ, Jan LY, Jan YN. Distinct morphogenetic functions of similar small GTPases: Drosophila Drac1 is involved in axonal outgrowth and myoblast fusion. Genes Dev. 1994;8:1787–1802. doi: 10.1101/gad.8.15.1787. [DOI] [PubMed] [Google Scholar]
- Nguyen HT, Bodmer R, Abmayr SM, McDermott JC, Spoerel NA. D-mef2: a Drosophila mesoderm-specific MADS box-containing gene with a biphasic expression profile during embryogenesis. Proc Natl Acad Sci U S A. 1994;91:7520–7524. doi: 10.1073/pnas.91.16.7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen HT, Xu X. Drosophila mef2 expression during mesoderm development is controlled by a complex array of cis-acting regulatory modules. Dev Biol. 1998;204:550–566. doi: 10.1006/dbio.1998.9081. [DOI] [PubMed] [Google Scholar]
- Nose A, Isshiki T, Takeichi M. Regional specification of muscle progenitors in Drosophila: the role of the msh homeobox gene. Development. 1998;125:215–223. doi: 10.1242/dev.125.2.215. [DOI] [PubMed] [Google Scholar]
- Ranganayakulu G, Zhao B, Dokidis A, Molkentin JD, Olson EN, Schulz RA. A series of mutations in the D-MEF2 transcription factor reveal multiple functions in larval and adult myogenesis in Drosophila. Dev Biol. 1995;171:169–181. doi: 10.1006/dbio.1995.1269. [DOI] [PubMed] [Google Scholar]
- Richardson B, Beckett K, Nowak S, Baylies MK. SCAR/WAVE and Arp2/3 are critical for cytoskeletal remodeling at the site of myoblast fusion. doi: 10.1242/dev.010678. In Revision. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Gomez M, Coutts N, Suster ML, Landgraf M, Bate M. myoblasts incompetent encodes a zinc finger transcription factor required to specify fusion-competent myoblasts in Drosophila. Development. 2002;129:133–141. doi: 10.1242/dev.129.1.133. [DOI] [PubMed] [Google Scholar]
- Ruiz-Gomez M, Romani S, Hartmann C, Jackle H, Bate M. Specific muscle identities are regulated by Kruppel during Drosophila embryogenesis. Development. 1997;124:3407–3414. doi: 10.1242/dev.124.17.3407. [DOI] [PubMed] [Google Scholar]


