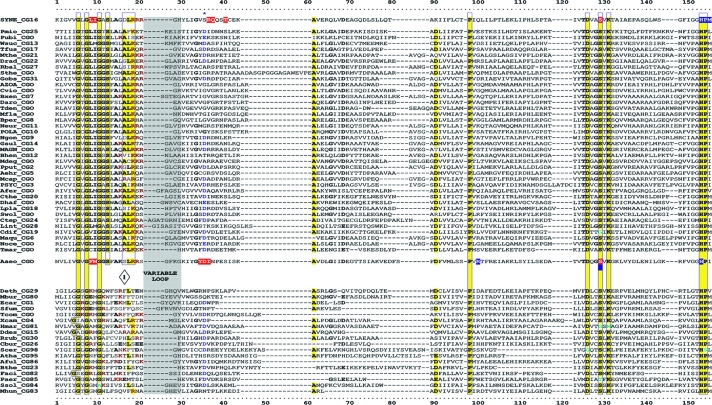
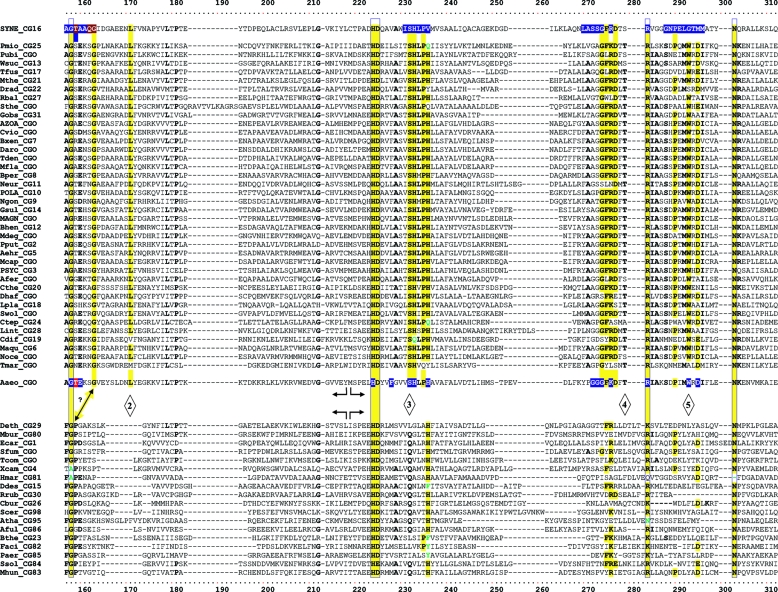
FIG. 3.
Master alignment of cohesion group representatives. The final manual alignment of 58 cohesion group representatives (see the appendix) was imported from the BioEdit alignment editor into the Word program to enhance presentation. TyrAα sequences are shown in the top section bounded at the top and bottom by sequences (Synechocystis sp. and Aquifex aeolicus) for which X-ray crystal structures are available. TyrAβ sequences are shown at the bottom. Amino acid residues shown to be important for NADP+ or for NAD+ in Synechocystis sp. and Aquifex aeolicus, respectively (48, 71), are shown in red with white lettering. Residues modeled in Synechocystis sp. and Aquifex aeolicus to be important for l-arogenate or for prephenate binding, respectively (48, 71), are shown in blue with white lettering. Relative residue position numbers are shown across the top. Invariant or near-invariant anchor residues are enclosed within vertical bars and highlighted yellow. Other highly conserved residues are shown in boldface type and highlighted yellow. Near-invariant residues that differ in a cohesion group representative, but which are nevertheless uniformly different throughout the cohesion group, are shown in boldface green type. The gray vertical band encloses residues in a variable loop (one to nine residues). Divergently pointed arrows at residue positions 216 and 217 mark the boundary between the pyridine nucleotide-binding domain and the catalytic domain. Regions that distinguish TyrAα and TyrAβ, as discussed in the text, are marked with numbers within triangles.