Abstract
Ehrlichia chaffeensis is an obligate intracellular bacterium and the causative agent of human monocytic ehrlichiosis. Although this pathogen grows in several mammalian cell lines, no general model for eukaryotic cellular requirements for bacteria replication has yet been proposed. We found that Drosophila S2 cells are permissive for the growth of E. chaffeensis. We saw morulae (aggregates of bacteria) by microscopy, detected the E. chaffeensis 16S rRNA gene by reverse transcriptase PCR, and used immunocytochemistry to detect E. chaffeensis in S2 and mammalian cells. Bacteria grown in S2 cells reinfected mammalian macrophages. S2 cells were made nonpermissive for E. chaffeensis through incubation with lipopolysaccharide. Our results demonstrate that S2 cells are an appropriate system for studying the pathogenesis of E. chaffeensis. The use of a Drosophila system has the potential to serve as a model system for studying Ehrlichia due to its completed genome, ease of genetic manipulation, and the availability of mutants.
Ehrlichia chaffeensis, the causative agent of human monocytic ehrlichiosis, was first reported in 1987. It has subsequently been reported in 30 U.S. states and was designated a nationally reportable disease by the U.S. Centers for Disease Control in 1999 (3). The bacteria are vectored by Amblyomma americanum (lone star tick), and white-tailed deer are considered to be the major reservoir for the bacteria. Although some details are known about the host response(s) of vertebrates to E. chaffeensis, little is known about the invertebrate response to the bacteria in the tick (7). Ehrlichiae are gram-negative, coccus (round)-shaped bacteria measuring 0.5 to 2 μm in diameter that, upon infection, form vacuole-bound colonies (called morulae) in leukocytes (2). In particular, E. chaffeensis infects mononuclear leukocytes (monocytes and macrophages). The clinical symptoms of the disease include fever, headache, chills, muscle aches, fatigue, nausea/vomiting, swollen glands, and/or delirium. Drugs such as tetracycline/doxycycline, which inhibit the protein synthesis of the bacteria by binding to the 30S ribosomal subunit, are often used during treatment(s) (2, 15). In approximately 40 to 60% of cases, hospitalization is necessary (20). Although the case fatality rates are approximately 3% (15), inappropriate treatment of the disease may lead to irreversible neurological damage due to the results of acute inflammatory responses.
Isolates of E. chaffeensis are most often cultured in the canine, macrophage-like cell line DH82. The bacteria also grow in other cell lines, including human microvascular endothelial cells, African green monkey kidney cells, human cervical epithelioid carcinoma cells, human monocytic leukemia cells, mouse embryo cells, buffalo green monkey cells, and murine fibroblasts (15). However, no clear requirements for cell tropism have been defined. The host genes that are required for the intracellular growth of E. chaffeensis are not known. Some efforts to characterize gene expression in mammalian macrophages have been initiated (21), but those studies have not revealed significant details about host requirements. The Drosophila melanogaster system offers several advantages over mammalian macrophages for these studies. For example, the D. melanogaster genome is well defined, with many of the innate immune response genes having homologues in mammals. S2 cells were chosen because E. chaffeensis is macrophage-tropic and S2 cells have characteristics of hemocytes, the insect equivalent of macrophages. S2 cells are more easily transfected than mammalian macrophages. This characteristic will be advantageous for experiments utilizing small interfering RNA techniques. Moreover, while the tick is the natural host for the bacteria, the tick system is less defined than that of D. melanogaster. Thus, the D. melanogaster system offers a closer parallel as an insect system versus a mammalian system for studying the tick-derived bacteria E. chaffeensis. Most recently, it has been recognized that insect systems are an extremely useful tool for studying the innate immune responses elicited by various pathogens in comparison to the response(s) observed in vertebrate systems (11). The D. melanogaster-derived cell line, S2 (Schneider line 2 cells), was isolated from primary cultures of 20- to 24-h-old embryos over 20 years ago (10). These cells are classified as hemocytes, which are known to circulate freely in the hemolymph of Drosophila and are phagocytic in nature. In addition, these cells are responsible for the synthesis/secretion of antimicrobial peptides (19). Indeed, Drosophila S2 cells have served as an in vitro model to study another obligate intracellular pathogen, Chlamydia trachomatis (8). The early steps of C. trachomatis infection in S2 cells mirror those seen in mammalian cells. Conservation of infection was observed between the steps of entry, inclusion formation, inhibition of phagolysosomal fusion, and acquisition of Golgi compartment-derived sphingolipids. Other intracellular bacteria, including Listeria monocytogenes and Mycobacteria marinum, have also been grown in S2 cells (14, 16, 18). Consequently, the use of S2 cells for the study of intracellular bacteria is helping to contribute to the elucidation of bacterial and cellular mechanisms that are important to these infections. To date, there are no reports of successful E. chaffeensis growth in Drosophila S2 cells. Therefore, we tested the hypothesis that E. chaffeensis would grow in D. melanogaster-derived S2 cells to determine if it could serve as a model system. The data in the manuscript document conditions that allow E. chaffeensis replication in S2 cells.
MATERIALS AND METHODS
Maintenance of cell lines and Ehrlichia chaffeensis infections.
The canine macrophage cell line DH82 was maintained at 37°C in Dulbecco's modified Eagle's medium with 3.5% fetal bovine serum and 3.5% Nu serum (DMEM-7). The E. chaffeensis Arkansas isolate was continuously cultivated in the DH82 cell line at 37°C, 8% CO2 in DMEM-7 medium. Bacteria were passaged when infectivity reached 80 to 90% as visualized by using cytospin-prepared slides (stained with Hema3 fixative and Dif-Quik stain) to monitor the formation of morulae in the cells. Infected cells were removed by scraping each plate with a cell scraper, transferring the culture to a conical tube, and vortexing the suspension with glass beads. The freed bacteria were purified by centrifuging the suspension at 600 × g for 20 min to remove cell debris. The bacteria-containing supernatant was removed, transferred to a sterile conical tube, and centrifuged at 15,000 × g for 20 min to pellet the free bacteria. The final supernatant was removed and discarded, and the pellet was resuspended in an appropriate amount of sterile phosphate-buffered saline (PBS). The purified bacteria were used to reinfect fresh DH82 cells. Drosophila S2 cells were cultivated at 28°C in Schneider's Drosophila medium (product no. 11720; Gibco, Grand Island, NY) supplemented with 10% fetal bovine serum (product no. S11150; Atlanta Biologicals).
Time course infections of DH82 and Drosophila S2 cells.
Drosophila S2 cells were plated in six-well tissue culture plates (60 mm, product no. 92406; Techno Plastic Products AG) at a concentration of 1 × 106 cells per ml. The cells were allowed to adhere for at least 30 min but no longer than 24 h. Bacteria purified from infected DH82 (85 to 100%) cells, approximately 3 × 107 bacteria, were added to S2 cell cultures and then were monitored for infection at 12, 24, 48, 72, 96, and 120 h postinfection (hpi). At each time point, RNA was isolated using TriReagent (product no. TR118; Molecular Research Center, Inc., Cincinnati, OH). S2 cells were removed by pipetting the cells off the dish and pelleted by centrifugation at 300 × g for 5 min. The supernatants were discarded, and 2 ml of TriReagent was used to lyse the pellet. For the DH82 cells, the spent medium was aspirated from the culture dishes and cells were removed from the dish using 2 ml of TriReagent and repeated pipetting. The TriReagent plus the cells was then transferred to polypropylene tubes, 200 μl of chloroform was added, and the mixtures were incubated on ice for 15 min and centrifuged for 15 min at 10,000 rpm. A 750-μl amount of the aqueous phase was mixed with 750 μl of isopropanol and incubated overnight at 4°C. Samples were then centrifuged for 15 min at 13,000 rpm (4°C). The pellets were washed with 75% ethanol, centrifuged for 10 min at 8,000 rpm (4°C), and resuspended in 50 μl of nuclease-free water. RNA concentrations were determined spectrophotometrically (NanoDrop Technologies, Wilmington, DE). Infections were quantitated by assessing the morula formation on cytospins of infected cells. Cells were randomly scored for the presence or absence of morulae. Lastly, the bacteria from infected S2 cells were isolated at each time point as described above. Purified bacteria (from each time point of the S2 time course) were used to reinfect fresh DH82 cells to determine if S2-grown bacteria would be infectious. These DH82 cells were set up as a time course experiment: cells were plated in six-well tissue culture plates at a concentration of 1 × 106 cells per ml and were subsequently infected with a range of concentrations of purified bacteria derived from S2 cells that had been infected for various lengths of time. The DH82 cells were assessed for infection 72 h later as described above. RNA was also isolated from the DH82 cells.
Assessment of bacterial numbers for infection.
The amount of bacteria used for infection experiments of DH82 and Drosophila S2 cells was estimated by using TaqMan-based real-time reverse transcriptase PCR (RT-PCR) as previously described (9, 17). This TaqMan-based assay targets the E. chaffeensis 16S rRNA gene. Real-time PCR was performed on 10-fold serial dilutions of RNA extracted from 80- to 100%-infected DH82 cells (three different samples) using a Smart Cycler system (Cepheid, Sunnyvale, CA). Standard curves were generated by plotting the log number of bacteria versus the corresponding threshold cycle value (average of the results of three experiments). The lowest detection limit, or the presence of 1 bacterium, was considered to be the dilution at which the threshold cycle value approaches 40 (0).
Determination of infection by RT-PCR.
Infections were assessed by RT-PCR, using a Promega access one-step RT-PCR kit (Madison, WI). Amounts of 500 to 1,000 ng of RNA were used for each reaction mixture. Each reaction mixture contained 1× buffer, 0.2 mM dNTPs, 2 μM forward primer, 2 μM reverse primer, 1.5 mM MgSO4, 1 U per μl DNA polymerase, 1 U per μl RT, and nuclease-free water for a final reaction mixture volume of 25 μl. RT-PCRs were performed in a (Eppendorf Mastercycler gradient) thermocycler, based on primers specific for the 16S rRNA gene of E. chaffeensis. The primers (Integrated DNA Technologies, Coralville, IA) used for detecting E. chaffeensis in both S2 and DH82 cells were RRG27 (5′ GTATTACCGCGGCTGCTGGCAC 3′) and RRG3 (5′ CAATTGCTTATAACCTTTTGGTTATAAAT 3′) (NCBI Core Nucleotide accession no. M73222) (4). The cycling conditions for these primers were as follows: 48°C for 5 min, 42°C for 5 min, 45°C for 5 min, 48°C for 30 min, 94°C for 4 min, and then 35 cycles of 94°C for 30 seconds, 52°C for 30 seconds, and 72°C for 1 min. The total RNA input was assessed by using housekeeping genes for ribosomal protein 49 (rp49) (NCBI Core Nucleotide accession no. U92431) (Drosophila) and canine glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (NCBI Core Nucleotide accession no. AB038240). The sequences for housekeeping primers included rp49 sequences (5′ATCGGTTACGGATCGAACAA 3′ and 5′GACAATCTCCTTGCGCTTCT 3′) for the Drosophila S2 cells and GAPDH sequences (5′GATTGTCAGCAATGCCTCCT 3′ and 5′GGCAGGTCAGATCCACAACT 3′) for the DH82 cells. The cycling conditions for the rp49 primers were as follows: 48°C for 45 min, 94°C for 2 min, and then 35 cycles of 94°C for 45 seconds, 50°C for 1 min, and 72°C for 1.5 min. The cycling conditions for the GAPDH primers were as follows: 48°C for 45 min, 94°C for 2 min, and then 35 cycles of 94°C for 30 seconds, 55°C for 1 min, and 68°C for 1 min. RT-PCRs were also performed without RT in order to assure that DNA was absent from the sample(s). RT-PCR products were identified on a ChemiImager after electrophoresis in 2% agarose gels and staining with ethidium bromide. The sized amplicons for each primer set were RRG 27 and 3 (430 bp), rp49 (165 bp), and GAPDH (308 bp).
Immunocytochemistry.
Specific E. chaffeensis infections were confirmed by immunocytochemical techniques. Immunocytochemistry was performed on E. chaffeensis-infected S2 cells, E. chaffeensis-infected DH82 cells, uninfected S2 cells, and uninfected DH82 cells. The cells were prepared using cytospin slides and dried for 24 h at room temperature. The cells were fixed with acetone, dried for 10 min, and outlined using a Pap Pen (The Binding Site, Inc., San Diego, CA). Samples were placed in PBS containing 0.01% Tween 20 for 5 min. The samples were blocked in 10% blocking solution (PBS containing 50% healthy goat serum) for 30 min at 37°C, washed in PBS-Tween 20 for 5 min, and incubated with the primary antibody (mouse anti-E. chaffeensis; 1:100 dilution) for 24 h in the dark at 4°C. The primary antibody was either healthy mouse serum or serum taken from mice infected with E. chaffeensis. After incubation, the slides were washed with PBS-Tween 20 for 5 min and were incubated for 1 h in the dark at room temperature with a 1:50 dilution of goat anti-mouse immunoglobulin G conjugated with rhodamine (Organon Teknika Corp., West Chester, PA). Samples were washed in PBS-Tween 20 for 5 min and then in distilled water for 5 min before viewing.
Activation of S2 cells with LPS.
To determine if lipopolysaccharide (LPS) activation would inhibit the growth of E. chaffeensis, S2 cells were plated at a concentration of 1 × 106 cells per plate in six-well plates and were allowed to adhere for at least 30 min. LPS (from Salmonella enterica serotype Minnesota; Sigma) was sonicated for 1 h and then added to each well at a concentration of 10 μg per ml. The cells plus LPS were incubated for 5 h. S2 cells were infected with E. chaffeensis purified from DH82 cells. RNA was extracted (using TriReagent) from each sample at 48 hpi and assessed for the presence of Ehrlichia cells by using RT-PCR, as previously described.
Transmission electron microscopy and sample preparation.
Infected S2 (72 hpi), uninfected S2, infected DH82 (72 hpi), and uninfected DH82 cells were pelleted (5- to 10-μl pellets) and immersion fixed in fixative containing 2% paraformaldehyde and 2% glutaraldehyde in 0.1 sodium cacodylate buffer (pH 7.4) for 16 h at room temperature with constant rotation. Each sample was washed three times for 5 min in 0.1 M sodium cacodylate buffer at room temperature with constant rotation. The samples were postfixed with 2% osmium tetroxide in 0.1 M sodium cacodylate buffer with constant rotation for 1 to 2 h and then washed three times for 5 min in 0.1 M sodium cacodylate buffer at room temperature with constant rotation. Samples were preembedded/stained with 2% uranyl acetate in 0.2 M sodium acetate buffer (pH 5.2) for 1 h at room temperature, light-protected, with constant rotation and subsequently washed three times for 5 min in 0.2 M sodium acetate buffer at room temperature with constant rotation. The samples were dehydrated in an ascending acetone series (50, 60, 70, 80, 90, 95, and 100%) with constant rotation at room temperature and then in 100% propylene oxide for 10 min. Infiltration of samples was accomplished by using EMBED 812/Araldite 502 resin at room temperature with constant rotation and consisted of 10 min in 1:1 propylene oxide:resin, 20 min in 1:2 propylene oxide:resin, 10 min in 100% resin. New 100% resin was finally added to the sample, allowed to infiltrate for 16 h, and then polymerized in a drying oven at 60°C for 24 to 48 h. The images were collected on a CM 100 (FEI Company) transmission electron microscope at 100 kV using a Hamamatsu C8484 digital camera and Advanced Microscopy Techniques Corp. software version 5.4.2.22B.
Statistics.
Statistical values were determined by using Student's t test (two-tailed, general). P values of <0.02 (as indicated) were considered highly significant. Data are presented as the mean ± standard deviation (SD). Differences were determined by using the StatMost statistical package (Data XIOM, Los Angeles, CA).
RESULTS
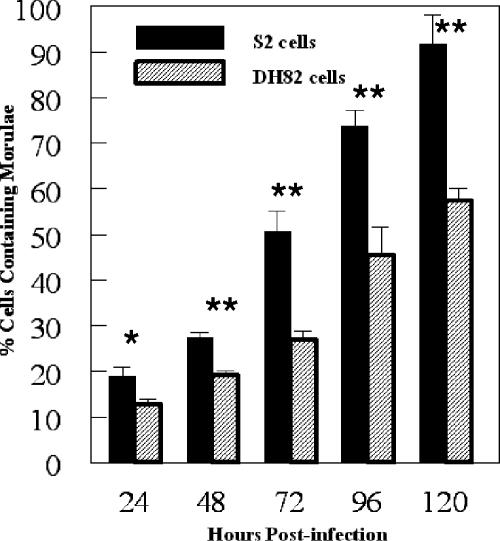
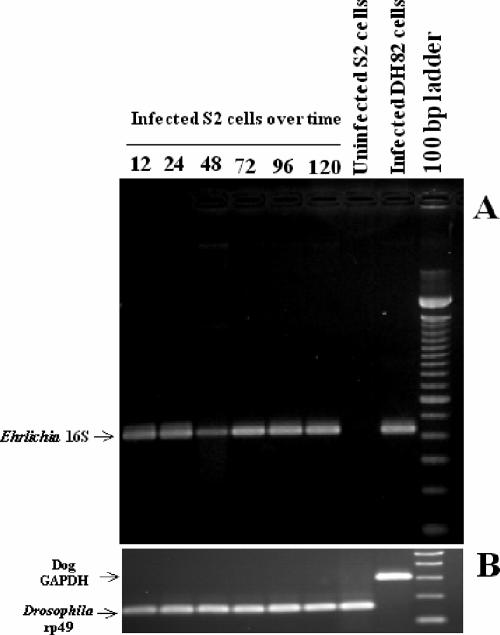
To test the hypothesis that E. chaffeensis can be propagated in macrophage-like S2 cells, we infected the S2 cells and assessed the infection via morphological, molecular, and immunological techniques. In our initial experiments, bacteria isolated from heavily infected DH82 cells (∼1.5 × 107 bacteria) were used in an attempt to infect the S2 cells. The S2 cells were tested for the presence of bacteria at time points ranging from 24 hpi to as late as 120 hpi. At 24 hpi, 18.8% of the S2 cells contained one or more morulae. At 120 hpi, 91.7% of the S2 cells contained one or more morulae (Fig. 1). We observed an increase in the percentage of cells infected with each subsequent time point. At the 120-h time point, many of the S2 cells contained large numbers of morulae. The S2 cells had more morulae than the DH82 cells at each time point beyond 24 hpi (P < 0.02) (Fig. 1). Moreover, the cells became progressively more vacuolated and misshapen throughout the infection. To confirm the infection in the S2 cells, RNA was assessed for the presence of the E. chaffeensis 16S ribosomal subunit transcript. Bacterial message was detected at 12 hpi through 120 hpi, which correlated with the observed morula formation in the infected S2 cells (Fig. 2). Additionally, we compared the kinetics of infection between S2 cells and DH82 cells. At 24 hpi, 12.6% of the DH82 cells contained one or more morulae, compared to almost 20% of the S2 cells. By 120 hpi, 57.4% of the DH82 cells contained morulae, compared to >90% of the S2 cells (Fig. 1). Therefore, it was clear that Ehrlichia bacteria were replicating in S2 cells.
FIG. 1.
Percentage of cells containing morulae in S2 and DH82 cells after infection with E. chaffeensis. Cells were considered positive when one or more morulae were present. Results are the averages of three separate infection experiments (mean ± SD). Asterisks show statistical significance: *, P < 0.06; **, P < 0.02.
FIG. 2.
(A) RT-PCR of E. chaffeensis 16S rRNA gene (band at 430 bp) at 12 to 120 hpi after infection of S2 cells. Time course infection experiments and RT-PCRs of 16S rRNA gene were repeated more than six times with the same outcome. The results of one representative experiment are shown. (B) RT-PCR of D. melanogaster rp49 (165 bp) and dog GAPDH (308 bp) housekeeping genes performed on samples shown in panel A.
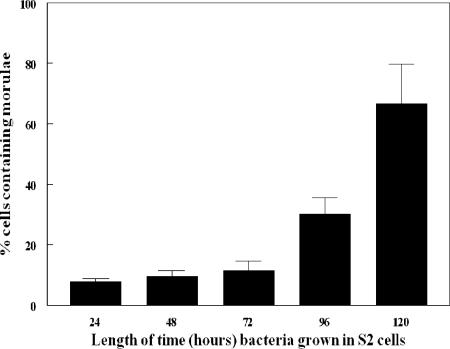
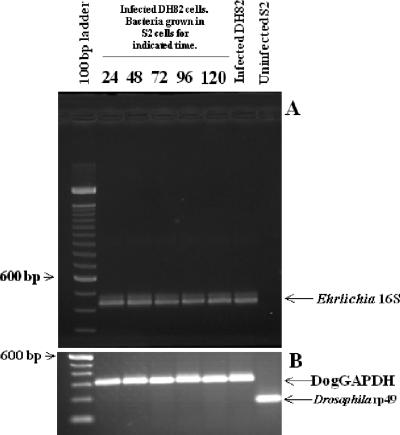
To confirm that the bacteria grown in S2 cells were completing replication and were infectious to vertebrate cells, DH82 cells were infected with bacteria purified from S2 cells at 24, 48, 72, 96, and 120 hpi of the S2 cells. The purified bacteria were used to infect the DH82 cells, and the amount of bacteria present in the DH82 cells was assessed 72 h later by quantitation of morulae. When S2 cells were only infected for 24 h, the bacteria isolated from those cells infected 8% of the DH82 cells (Fig. 3). Sixty-seven percent of the DH82 cells were infected with bacteria isolated from S2 cells infected for 120 h (Fig. 3). We confirmed these infections by RT-PCR of the 16S E. chaffeensis rRNA (Fig. 4). Consequently, E. chaffeensis is capable of completing its life cycle in S2 cells and also maintains its ability to reinfect the mammalian macrophage cell line DH82.
FIG. 3.
Growth of E. chaffeensis (originally grown in S2 cells) in DH82 cells. Bacteria grown in S2 cells for the indicated length of time were used to infect DH82 cells. The percentage of DH82 cells containing morulae was assessed 72 h later. Results are the averages of three separate time course experiments (mean ± SD).
FIG. 4.
(A) Reinfection of DH82 cells with S2-cell-grown bacteria. RT-PCR of E. chaffeensis 16S rRNA gene (band at 430 bp) after DH82 cells were infected with bacteria grown in S2 cells for the indicated times. (B) RT-PCR of D. melanogaster rp49 (165 bp) and dog GAPDH (308 bp) housekeeping genes performed on samples shown in panel A.
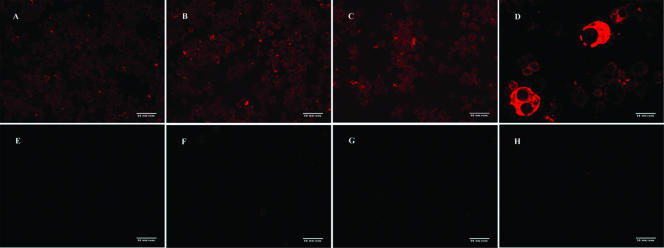
To further demonstrate that the inclusions seen in the S2 cells after infection with E. chaffeensis were bacteria, we utilized immunocytochemistry to detect bacteria. Slides were made with infected S2 and DH82 cells or with uninfected cells. Sera collected from E. chaffeensis-infected mice or from healthy mice were used as primary antibodies. Only E. chaffeensis-specific antiserum reacted with the infected DH82 (Fig. 5D) and S2 cells (Fig. 5A to C). No bacteria were detected with the healthy mouse serum incubated with infected or uninfected cells or with secondary antibody alone (Fig. 5E to H).
FIG. 5.
Immunocytochemical detection of Ehrlichia in S2 and DH82 cells. Shown are infected S2 cells incubated with E. chaffeensis-specific mouse serum and secondary antibody (A, B, C), infected DH82 cells incubated with E. chaffeensis-specific mouse serum and secondary antibody (D), infected S2 cells incubated with healthy mouse serum and secondary antibody (E), infected S2 cells incubated with secondary antibody only (F), uninfected S2 cells incubated with E. chaffeensis-specific mouse serum and secondary antibody (G), and uninfected DH82 cells incubated with E. chaffeensis-specific mouse serum and secondary antibody (H). Each image was captured at ×20 magnification.
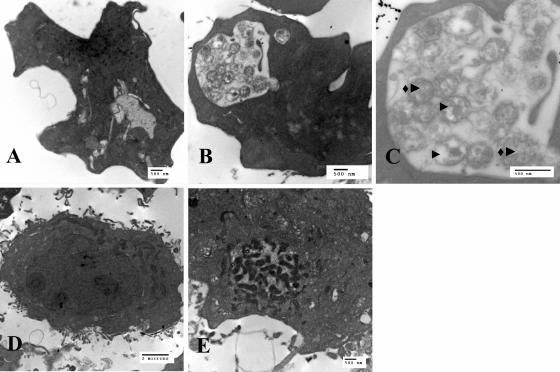
We also demonstrated the presence of morulae in S2 and DH82 cells by using transmission electron microscopy (Fig. 6). The S2 and DH82 cells used in this experiment were infected at the same time with the same batch of E. chaffeensis and were both fixed and imaged at 72 hpi. Morulae were seen in both S2 and DH82 cells. By 72 hpi, most of the morulae in the DH82 cells contained numerous dense, elongated bacterial forms (Fig. 6E). The morulae in the S2 cells also contained many bacteria, with both reticulate and dense forms (Fig. 6B and C). The morulae in both cell types were confined to vacuoles. Uninfected S2 and DH82 cells contained minimal vacuoles. In instances where vacuoles were seen in the uninfected cells (Fig. 6A), they contained no bacteria.
FIG. 6.
Transmission electron micrographs of uninfected or E. chaffeensis-infected S2 and DH82 cells. Shown are uninfected S2 cells (A), infected S2 cells (B), infected S2 cells (black arrowheads indicate dense core form and diamond with black arrowhead indicates reticulate form) (C), uninfected DH82 cells (D), and infected DH82 cells (E).
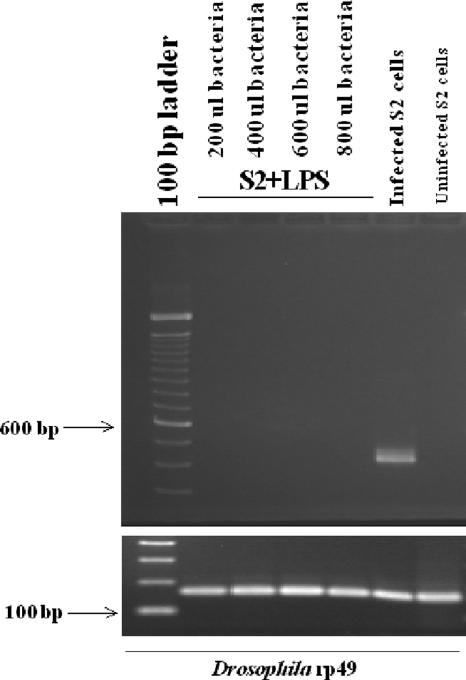
Lastly, it is known that mammalian macrophages are nonpermissive for E. chaffeensis growth after LPS activation (13). Therefore, we wanted to determine if S2 cells could be resistant to E. chaffeensis infection. To test this hypothesis, we incubated S2 cells with LPS (10 μg per ml) for 4 h, and then we challenged the cells with increasing doses of purified E. chaffeensis bacteria. RNA was extracted from the cells at 48 hpi, and the RNA samples were analyzed by RT-PCR for Ehrlichia infection. S2 cells activated with LPS (10 μg per ml) were nonpermissive for E. chaffeensis regardless of the number of bacteria used for infection (Fig. 7). As a control, unstimulated S2 cells were infected at the same time using the same number of bacteria (Fig. 7). Thus, activated S2 cells become nonpermissive to infection by E. chaffeensis, similar to results seen in activated human monocytes (13).
FIG. 7.
RT-PCR results following LPS activation of S2 cells. E. chaffeensis 16S rRNA (band at 430 bp) is present only in cells not treated with LPS. S2 cells were infected with increasing numbers of bacteria at 1.4 × 107 bacteria/200 μl. Housekeeping gene is Drosophila melanogaster rp49. ul, μl.
DISCUSSION
We have presented novel data showing that Drosophila S2 cells can be successfully infected with the obligate, intracellular bacterium E. chaffeensis. Infection was confirmed using three distinct criteria: (i) the presence of morulae in the S2 cells; (ii) the detection of 16S rRNA of E. chaffeensis in infected cells; and (iii) the specific detection of E. chaffeensis in S2 cells by a bacterium-specific antibody. Morulae in mammalian cells are known to have two forms: dense-core and reticulate cells (15). We found the presence of both dense and reticulate forms of bacteria in S2 cells with, particularly, a preponderance of reticulate forms in infected S2 cells. Considering the differences observed between bacterial growth in the S2 and DH82 cells (Fig. 1), this is not surprising. The reticulate form is the dividing form of the bacteria, and thus, an increased number of dividing bacteria coincides with the increased bacterial kinetics observed in the S2 cells compared to the kinetics in the DH82 cells. Zhang et al. (20) found that the dense-core form of the bacteria was present exclusively during early infection (0 to 1 hpi). Between 24 and 48 hpi, the reticulate form predominated and binary fission was often observed. Subsequent to that, dense-core forms reappeared. It is possible that the reticulate forms predominate in insect cells grown at lower temperatures or that the reappearance of dense-core forms occurs later. Alternatively, the presence of reticulate and dense-core forms may be cell line and/or temperature dependent. Additional electron microscopy work will be necessary to determine this. Nevertheless, these data confirm that E. chaffeensis can be propagated in S2 cells and that its developmental patterns are similar to those seen in mammalian macrophages. In addition, vacuoles containing appropriately sized inclusions (presumably bacteria) were observed by light microscopy/live video imaging in both S2 cells and DH82 cells that had been infected with E. chaffeensis (data not shown). The bacteria were confined to the vacuoles in the infected cells; conversely, no bacteria were observed in the uninfected S2 or DH82 cells.
The kinetics of bacterial infection of S2 cells was comparable to the growth of bacteria in DH82 cells, which are one of the cell lines most commonly used to propagate infections of E. chaffeensis. Moreover, bacteria isolated from infected S2 cells were infectious and could be used to infect DH82 cells. Therefore, the growth of E. chaffeensis in dipteran insect cells does not compromise the viability of the bacteria. This is consistent with the natural life cycle of Ehrlichia that includes an arachnid host, the tick. The host requirements for E. chaffeensis to grow in ticks are not clear. The D. melanogaster system offers the advantage of having a well-defined genome. Using the S2 cells to identify which host genes are necessary to support E. chaffeensis growth will eventually allow us to study homologous genes in both arachnids and vertebrates. Our data are also consistent with observations that S2 cells are capable of supporting other macrophage-tropic intracellular bacteria. For example, Chlamydia trachomatis, Listeria monocytogenes, and Mycobacterium fortuitum all infect S2 cells (1, 5, 6, 8, 14, 16). Studies of C. trachomatis bacteria in S2 cells have provided valuable information about its early and late infection processes (8). S2 cells have also been critical to the identification of key components involved in the pathogenesis of L. monocytogenes (6) and the involvement of CD36 in M. fortuitum infections (16). Nevertheless, our findings open the molecular and genetic tool box of the Drosophila genome for the study of E. chaffeensis.
In addition to the observation that E. chaffeensis is capable of infecting Drosophila S2 cells, we also were able to make the S2 cells nonpermissive for infection by E. chaffeensis. Although E. chaffeensis does not make LPS or peptidoglycan (12), we wanted to determine if LPS could activate the S2 cells. Activation with LPS prevented bacterial replication even at high multiplicities of infection. More importantly, the option of a system made nonpermissive by the addition of LPS increases the utility of S2 cells and makes them comparable to vertebrate macrophages in this respect (13). In particular, comparing gene expression levels in activated versus infected S2 cells will allow us to pinpoint genes specific to E. chaffeensis infections, as well as genes that are simply activation specific. It is our goal to define the mechanisms that allow for the maintenance of E. chaffeensis in its invertebrate and vertebrate hosts.
Acknowledgments
This study was supported by National Institutes of Health grants AI052206, AI55052, RR16475, and RR17686; NASA grants NAG2-1274 and NCC-8-242; and Kansas Agricultural Experiment Station Animal Health Project grant 481848.
This is Kansas Agriculture Experiment state publication 07-271-J.
Footnotes
Published ahead of print on 1 February 2008.
REFERENCES
- 1.Agaisse, H., L. S. Burrack, J. A. Philips, E. J. Rubin, N. Perrimon, and D. E. Higgins. 2005. Genome-wide RNAi screen for host factors required for intracellular bacterial infection. Science 309:1248-1251. [DOI] [PubMed] [Google Scholar]
- 2.Bakken, J. S., and J. S. Dumler. 2000. Human granulocytic ehrlichiosis. Clin. Infect. Dis. 31:554-560. [DOI] [PubMed] [Google Scholar]
- 3.Cao, W. C., Y. M. Gao, P. H. Zhang, X. T. Zhang, Q. H. Dai, J. S. Dumler, L. Q. Fang, and H. Yang. 2000. Identification of Ehrlichia chaffeensis by nested PCR in ticks from southern China. J. Clin. Microbiol. 38:2778-2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheng, C., C. D. Paddock, and R. Reddy Ganta. 2003. Molecular heterogeneity of Ehrlichia chaffeensis isolates determined by sequence analysis of the 28-kilodalton outer membrane protein genes and other regions of the genome. Infect. Immun. 71:187-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheng, L. W., and D. A. Portnoy. 2003. Drosophila S2 cells: an alternative infection model for Listeria monocytogenes. Cell. Microbiol. 5:875-885. [DOI] [PubMed] [Google Scholar]
- 6.Cheng, L. W., J. P. Viala, N. Stuurman, U. Wiedemann, R. D. Vale, and D. A. Portnoy. 2005. Use of RNA interference in Drosophila S2 cells to identify host pathways controlling compartmentalization of an intracellular pathogen. Proc. Natl. Acad. Sci. USA 102:13646-13651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Childs, J. E., and C. D. Paddock. 2003. The ascendancy of Amblyomma americanum as a vector of pathogens affecting humans in the United States. Annu. Rev. Entomol. 48:307-337. [DOI] [PubMed] [Google Scholar]
- 8.Elwell, C., and J. N. Engel. 2005. Drosophila melanogaster S2 cells: a model system to study Chlamydia interaction with host cells. Cell. Microbiol. 7:725-739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ganta, R. R., C. Cheng, E. C. Miller, B. L. McGuire, L. Peddireddi, K. R. Sirigireddy, and S. K. Chapes. 2007. Differential clearance and immune responses to tick cell-derived versus macrophage culture-derived Ehrlichia chaffeensis in mice. Infect. Immun. 75:135-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Han, K. 1996. An efficient DDAB-mediated transfection of Drosophila S2 cells. Nucleic Acids Res. 24:4362-4363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kavanagh, K., and E. P. Reeves. 2007. Insect and mammalian innate immune responses are much alike. Microbe 2:596-599. [Google Scholar]
- 12.Lin, M., and Y. Rikihisa. 2003. Ehrlichia chaffeensis and Anaplasma phagocytophilum lack genes for lipid A biosynthesis and incorporate cholesterol for their survival. Infect. Immun. 71:5324-5331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin, M., and Y. Rikihisa. 2004. Ehrlichia chaffeensis downregulates surface Toll-like receptors 2/4, CD14 and transcription factors PU.1 and inhibits lipopolysaccharide activation of NF-kappa B, ERK 1/2 and p38 MAPK in host monocytes. Cell. Microbiol. 6:175-186. [DOI] [PubMed] [Google Scholar]
- 14.Mansfield, B. E., M. S. Dionne, D. S. Schneider, and N. E. Freitag. 2003. Exploration of host-pathogen interactions using Listeria monocytogenes and Drosophila melanogaster. Cell. Microbiol. 5:901-911. [DOI] [PubMed] [Google Scholar]
- 15.Paddock, C. D., and J. E. Childs. 2003. Ehrlichia chaffeensis: a prototypical emerging pathogen. Clin. Microbiol. Rev. 16:37-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Philips, J. A., E. J. Rubin, and N. Perrimon. 2005. Drosophila RNAi screen reveals CD36 family member required for mycobacterial infection. Science 309:1251-1253. [DOI] [PubMed] [Google Scholar]
- 17.Sirigireddy, K. R., and R. R. Ganta. 2005. Multiplex detection of Ehrlichia and Anaplasma species pathogens in peripheral blood by real-time reverse transcriptase-polymerase chain reaction. J. Mol. Diagn. 7:308-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stroschein-Stevenson, S. L., E. Foley, P. H. O'Farrell, and A. D. Johnson. 2006. Identification of Drosophila gene products required for phagocytosis of Candida albicans. PLoS Biol. 4:e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vilmos, P., and E. Kurucz. 1998. Insect immunity: evolutionary roots of the mammalian innate immune system. Immunol. Lett. 62:59-66. [DOI] [PubMed] [Google Scholar]
- 20.Zhang, J. Z., V. L. Popov, S. Gao, D. H. Walker, and X. J. Yu. 2007. The developmental cycle of Ehrlichia chaffeensis in vertebrate cells. Cell. Microbiol. 9:610-618. [DOI] [PubMed] [Google Scholar]
- 21.Zhang, J. Z., M. Sinha, B. A. Luxon, and X. J. Yu. 2004. Survival strategy of obligately intracellular Ehrlichia chaffeensis: novel modulation of immune response and host cell cycles. Infect. Immun. 72:498-507. [DOI] [PMC free article] [PubMed] [Google Scholar]