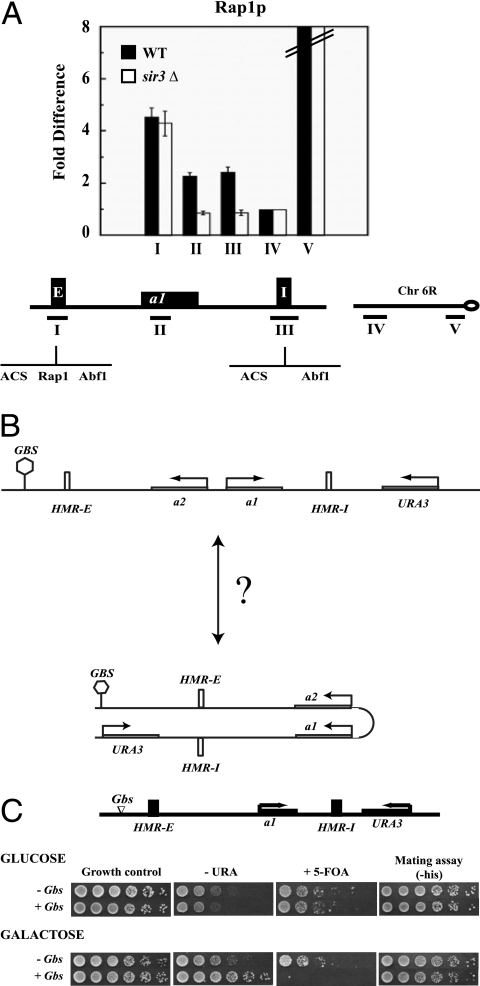
FIG. 3.
Functional long-range communication at HMR. (A) Mapping Rap1p at HMR. Antibodies against the Rap1p C terminus were used to map the distribution of Rap1p across the HMR locus in a wild-type (WT) strain and a sir3Δ strain. Quantitative ChIPs were performed as described for Fig. 2. The PCR probes used are shown in the schematic diagram. Error bars indicate standard errors. (B) Schematic representation of the enhancer construct at HMR. The locations of Gal4p binding sites and the URA3 gene are shown. (C) “Enhancer” activity at HMR. Strains with URA3 located downstream of HMR-I containing no Gal4 binding sites (-Gbs) or four Gbs upstream of HMR-E (+Gbs), were grown in 5 ml YPD overnight. Cells were washed, and fivefold serial dilutions were prepared. Properly supplemented YM plates containing 2% galactose (YMG) or 2% glucose (YMD) as a carbon source were used to induce or to repress expression of Gal4p, respectively. To assay for expression of URA3, cells were spotted onto YMG or YMD plates lacking or containing uracil or 5-FOA and photographed. To assay for expression of MATa1 at HMR, cells were spotted onto properly supplemented YMG or YMD plates with mating lawns.