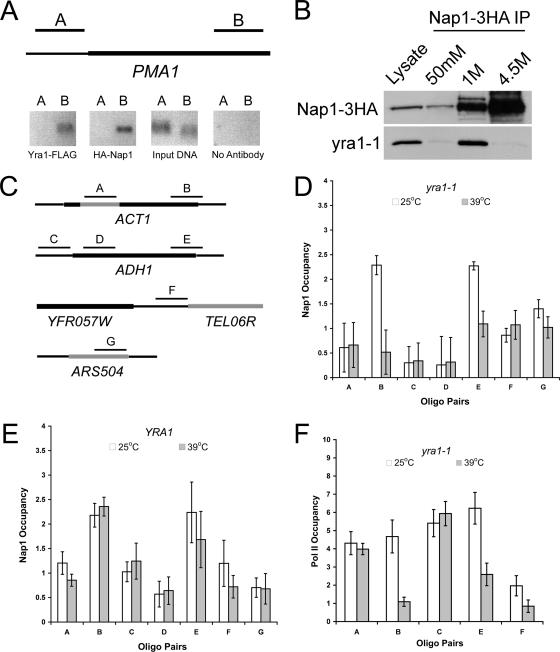
FIG. 5.
Nap1 recruitment to sites of active transcription is Yra1 dependent. (A) Chromatin was isolated from a Δyra1 Δnap1 strain expressing plasmids encoding Yra1-FLAG and HA-Nap1. Yra1 and Nap1 occupancy at the PMA1 promoter and ORF was determined by ChIP assay with conventional PCR using oligonucleotide pairs to the indicated regions of PMA1. The assay was also performed without antibody as a negative control. (B) yra1-1 was coimmunoprecipitated from whole-cell extracts with Nap1-3HA and detected by Western blotting. (C) Nap1 recruitment to the ACT1 and ADH1 ORFs and intergenic regions was tested by ChIP assay using the indicated oligonucleotide pairs. (D) Nap1 ChIP assays quantified by real-time PCR were performed with a Δyra1 Δnap1 strain expressing yra1-1 and HA-Nap1. Cells were grown for 2 h at 25°C or 39°C prior to fixation as indicated. Occupancy is expressed as the log2 difference (n-fold) between specific and background IP signals normalized to input, displayed as the mean and average standard deviation from two independent experiments. Oligo, oligonucleotide. (E) Nap1 ChIP assays quantified by real-time PCR were performed exactly as for panel D with a YRA1 Δnap1 strain expressing HA-Nap1. (F) RNAP II (Pol II) ChIP assays quantified by real-time PCR as for panel D were performed from a Δyra1 strain expressing yra1-1 exogenously. Means and standard deviations are indicated.