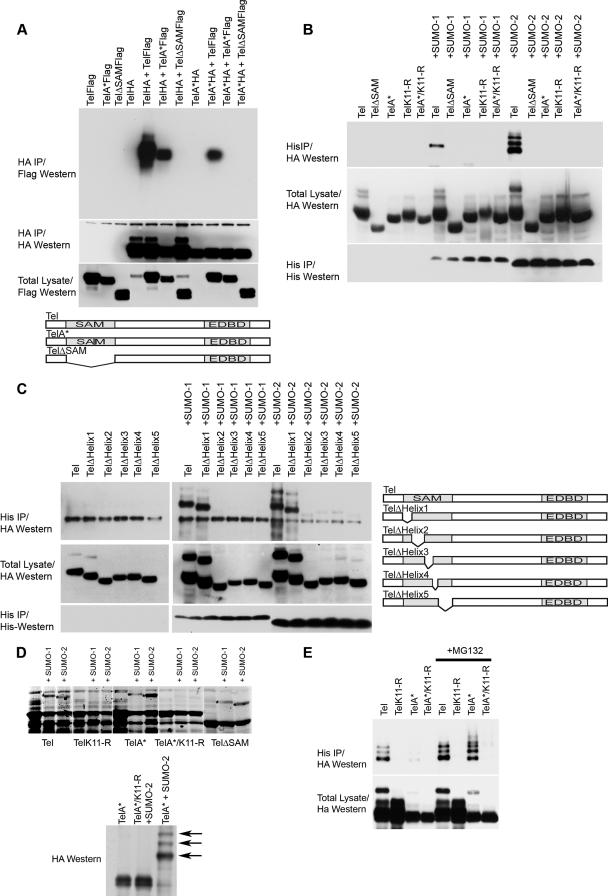
FIG. 2.
K11 of Tel polymers, but not Tel monomers, is found conjugated with SUMO in cells. (A) Disruption of the SAM domain prevents Tel self-association. HA epitope-tagged and Flag epitope-tagged fusions of wild-type Tel or Tel expressing mutations that disrupt the SAM domain (A*, which contains an arginine residue in place of an alanine residue; deletion of the SAM domain is shown as ΔSAM) were expressed in 293T cells in the indicated combinations. A schematic representation of the mutants tested is included. (B and C) Monomeric forms of Tel exhibit low levels of sumoylation in cells. (B) HA epitope-tagged versions of wild-type Tel or Tel expressing mutations that disrupt the SAM domain (described above) were cotransfected into 293T cells along with His epitope-tagged SUMO-1 or SUMO-2, and a sumoylation assay was performed. (C) The experiment in panel B was performed using Tel mutants in which each of the five helices that comprise the SAM domain were individually deleted. A schematic representation of the mutants tested is included. (D) Tel monomers are efficiently sumoylated in vitro on K11. Fusions between GST and either full-length wild-type Tel or full-length Tel harboring mutations that disrupt the SAM domain (see above) were sumoylated in E. coli as described in the legend of Fig. 1A. Sumoylated forms of Tel are highlighted (*) and were confirmed by Western blotting and also MS (data not shown). Shown also is an HA Western blot to detect Tel following an in vitro sumoylation assay (37) using the indicated Tel proteins that were made by in vitro translation in the presence of 1 mM unlabeled methionine. Sumoylated proteins that are absent from TelA* proteins containing a mutation of K11 to an arginine (TelA*/K11-R) are highlighted with arrows. (E) Inhibiting proteasome function stabilizes the pool of Tel monomers sumoylated on K11 in cells. Cells were cotransfected with the indicated Tel constructs along with His epitope-tagged SUMO-2. Following incubation with or without MG132, a sumoylation assay was performed. IP, immunoprecipitation.