FIG. 5.
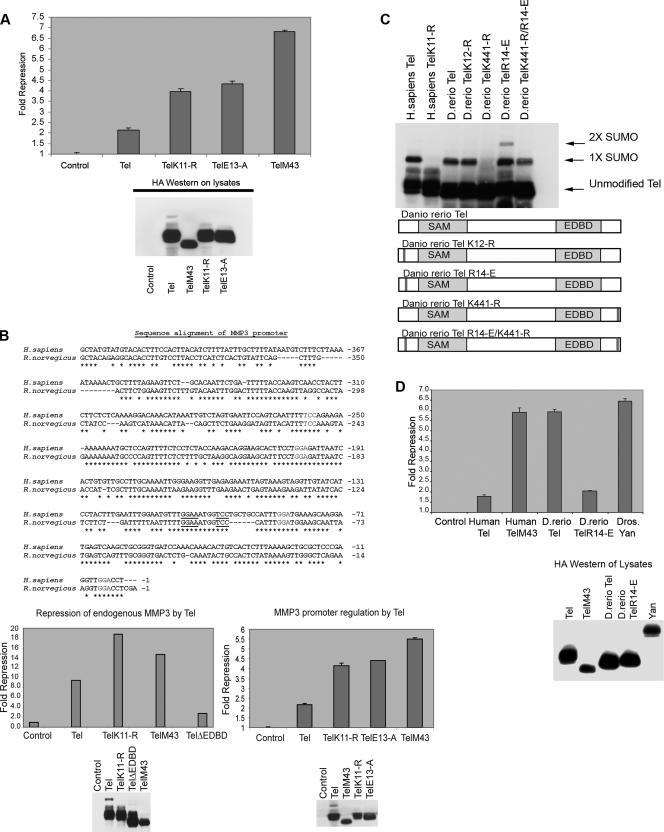
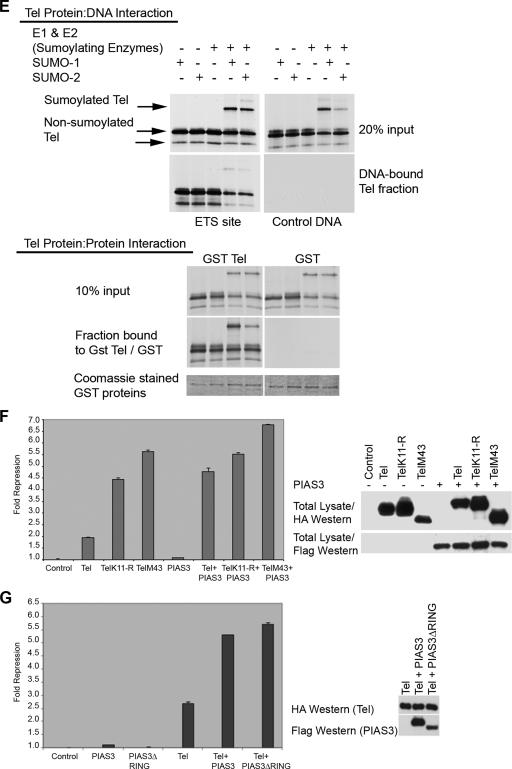
Sumoylation of K11 inhibits repression by Tel. (A) Either 293T, U2OS, or NIH 3T3 cells were cotransfected with the indicated Tel constructs along with a luciferase reporter in which an engineered minimal Tel/Yan regulatable promoter controls TK-driven luciferase expression. Equivalent transfection efficiencies of the indicated constructs were determined by a β-galactosidase assay for expression of a cotransfected LacZ reporter, and protein levels of ectopically expressed Tel constructs were determined by Western blotting of lysates. Shown is a representative experiment using 293T cells. Experiments were performed in triplicate. (B) Tel repression of expression of endogenous human stromelysin-1 (MMP3) is inhibited by sumoylation of K11. The upper panel shows that the promoters of rat and human stromelysin-1 share a number of conserved, potential Tel DNA binding sites. Highlighted in bold are previously characterized Tel binding sites (5). Additional highly conserved putative Tel binding sites are underlined. In the lower left panel, we established stable cell lines expressing the indicated Tel constructs. We prepared cDNA from these lines and performed quantitative PCR to assess the levels of expression of endogenous stromelysin-1. Shown graphically are the relative repression levels normalized against a control human gapdh gene. Shown in the right panel is a luciferase reporter assay. Luciferase expression was placed under the control of the human stromelysin promoter. Equivalent transfection efficiencies of the indicated constructs were determined by a β-galactosidase assay for expression of a cotransfected LacZ reporter, and protein levels of ectopically expressed Tel constructs were determined by Western blotting of lysates. Shown is a representative experiment using U2OS cells. Experiments were performed in triplicate. (C) D. rerio Tel does not express an N-terminal site of sumoylation but is sumoylated at position K441. Cells were transfected with the indicated constructs together with SUMO-2, and Tel sumoylation was assessed by Western blotting. D. rerio TelK12-R is the same as wild-type D. rerio Tel except that the lysine residue at position 12 has been replaced with an arginine residue. D. rerio TelR14-E is identical to wild-type D. rerio Tel except that the N-terminal sumoylation consensus site has been created by replacing the arginine residue with a glutamic acid residue at position 14. D. rerio TelK441-R is the same as wild-type D. rerio Tel except that the lysine residue at position 441 has been replaced with an arginine residue. D. rerio TelK441-R/R14-E is the same as D. rerio TelK441-R except that the arginine residue at position 14 has been replaced with a glutamic acid residue. A schematic representation of the mutants tested is included. (D) Cells were transfected with the indicated constructs, and a luciferase repression assay was performed as described in panel A. (E) Sumoylation of K11 impedes the ability of Tel to associate with DNA. Tel proteins were efficiently sumoylated following in vitro translation in reticulocyte lysates as previously described (37). Biotinylated Ets DNA-binding sites were coupled to streptavidin beads following the manufacturer's advice (Invitrogen) and incubated with the indicated proteins. Associated Tel proteins were recovered by SDS-PAGE. Tel-Tel interactions were assessed by incubating GST Tel fusion proteins or GST proteins alone, along with sumoylated and nonsumoylated in vitro translated forms of Tel. (F and G) PIAS3 enhancement of repression by Tel is sumoylation independent. The experiment shown in panel F is the same as that described for panel A except that cells were cotransfected with or without PIAS3. In the experiment in panel G, cells were transfected with the indicated constructs, and a luciferase reporter assay was performed as described for panel A. PIAS3ΔRING is the same as PIAS3 except that the RING domain has been deleted.