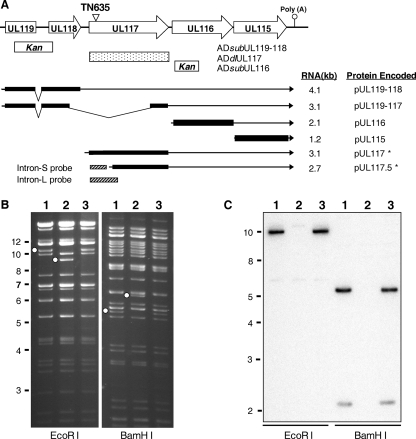
FIG. 1.
Construction of UL119-UL117 recombinant BAC-HCMV clones. (A) Viral genomic region encoding UL119-UL115. The first line represents the schematic structure of the viral genomic region. Viral ORFs are indicated by boxed arrows. Also indicated are the locations of the poly(A) signal downstream of UL115 and the transposon insertion in the recombinant BAC-HCMV clone TN635 (termed pADinUL117 in this study). The boxes below the first line represent the locations of the viral sequences within the UL119-UL116 region that are deleted in the substitution or deletion mutants, as indicated. Previously reported transcripts derived from the UL119-UL115 region (2, 23, 39) and the UL117-specific transcripts identified in this study (indicated by asterisks) are shown as lines with arrows representing the 3′ ends of the transcripts. Also indicated are the positions of two DNA probes used in Northern blotting analysis. Kan, kanamycin resistance gene cassette. (B) EcoRI and BamHI restriction digestion analysis and (C) Southern blotting analysis of UL117 recombinant BAC-HCMV clones. Open dots indicate restriction fragments unique to a recombinant BAC clone due to engineered sequence alteration. For Southern blotting analysis, a 32P-labeled probe of the UL117 ORF was used to hybridize EcoRI- or BamHI-digested BAC-HCMV DNA. Markers of molecular size (in kb) are indicated. Lane 1, the wild-type BAC pAD-GFP; lane 2, the UL117-deletion mutant BAC pADdlUL117; lane 3, the marker-rescued BAC pADrevUL117-1.