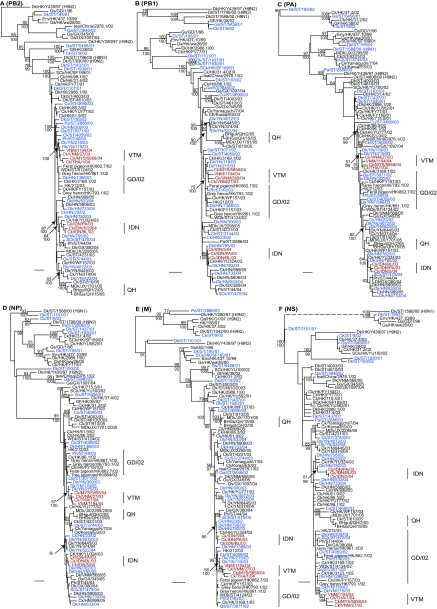
FIG. 3.
Phylogenetic relationships of the PB2 (A), PB1 (B), PA (C), NP (D), M (E), and NS (F) genes of representative influenza A viruses. The numbers above and below the branch nodes indicate neighbor-joining bootstrap values of ≥50% and Bayesian posterior probabilities of >95, respectively. Analyses were based on nucleotides 986 to 2288, 1 to 1480, 1404 to 2151, 1 to 990, 1 to 998, and 1 to 842, respectively. All gene trees were rooted to A/equine/Prague/1/56 except PB1 and PA, which were midpoint rooted. Colors indicate viruses isolated from southern China (blue), Vietnam (red), and Indonesia (red). Scale bar, 0.01 nucleotide substitutions per site. Abbreviations: BHGs, bar-headed goose; BHgull, brown-headed gull; Ck, chicken; Cu, chukkar; Dk, duck; FJ, Fujian; Gf, Guinea fowl; Gs, goose; GD, Guangdong; GX, Guangxi; GY, Guiyang; HK, Hong Kong; HN, Hunan; IDN, Indonesia; JX, Jiangxi; Mall, mallard; MDk, migratory duck; MYS, Malaysia; NGA, Nigeria; Pa, partridge; Ph, pheasant; Qa, quail; QH, Qinghai; SCk, silky chicken; ST, Shantou; THA, Thailand; VNM, Vietnam; WDk, wild duck; YN, Yunnan.