Fig. 3.
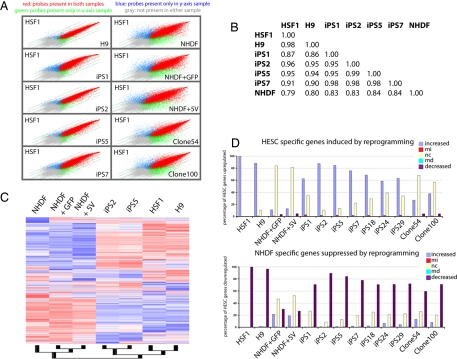
The transcriptome of iPS clones is highly similar to that of HESC. (A) Scatter-plot presentation of the expression values for all probe sets derived from genome-wide microarray expression data of indicated cell types. NHDF1 + GFP and NHDF1 + 5V denote a pool of fibroblasts infected with pMX/pMX-GFP control viruses or viruses carrying the five defined factors plus GFP at day 18 after infection. Like the H9 HESC line, iPS clones 2 and 5 appear highly similar to the HSF1 HESC, whereas iPS lines 1 and 7 appear slightly less similar to HESC. (B) Global Pearson correlation of the entire expression data (from Affymetrix microarrays) between indicated cell types. (C) Hierarchical clustering of gene-expression data of the indicated cell types. Normalization and expression analysis was performed with DNA-chip analyzer (dChip). A 20% presence call was used to filter genes for clustering, and redundant probe sets were removed. (D) The 2,000 most up- and down-regulated genes in HSF1 versus NHDF were determined from genome-wide expression datasets and analyzed for up-regulation, down-regulation, or no change in expression between iPS clones or pools of infected NHDF cells and NHDF. MI and MD denote statistically marginal increase or decrease, respectively.