Abstract
We examined the sera of six patients before and after i.v. infusions of autologous chronic lymphocytic leukemia (CLL) cells transduced ex vivo with an adenovirus encoding CD154 (Ad-CD154). Five patients made high-titer antibodies against adenovirus and three made IgG reactive with a leukemia-associated surface antigen, which we identified as ROR1. Anti-ROR1 antibodies were not detected in the sera of untreated patients. We generated anti-ROR1 mAbs and found they reacted specifically with the CLL cells of all patients, but not with nonleukemic leukocytes, a wide variety of normal adult tissues, or blood mononuclear cells, including CD5+ B cells of healthy adults. ROR1 could bind Wnt5a, which induced activation of NF-κB when coexpressed with ROR1 in HEK293 cells and enhanced the survival of CLL cells in vitro, an effect that could be neutralized by posttreatment anti-ROR1 antisera. We conclude that patients with CLL can break immune tolerance to ROR1, which is an oncofetal surface antigen and survival-signaling receptor in this neoplastic disease.
Keywords: chronic lymphocytic leukemia, neoplasia
Patients with chronic lymphocytic leukemia (CLL) typically develop hypogammmaglobulinemia and progressive immune deficiency, which impairs their immune response to vaccines (1–3). Implicated in the abnormal immune function are immune-suppressive factors (4, 5) and an acquired functional deficiency of the CD40 ligand (CD154) (6). Furthermore, CLL cells are particularly inept at antigen presentation, which appears in part secondary to inadequate leukemia cell expression of immune costimulatory and adhesion molecules (7–9).
Activation of CLL cells via CD40 ligation can reverse this immune-suppressive phenotype (7, 8, 10). Although CLL cells express class I and II major histocompatibility complex antigens, CD54 (ICAM-1), CD27, and CD40, these cells have low-to-absent expression of important immune costimulatory molecules, such as CD80, and cannot stimulate even allogeneic T cells in mixed lymphocyte reactions. However, upon interaction with cells that express CD154, CLL cells are induced to express immune costimulatory molecules, allowing them to become effective antigen-presenting cells (7). Similarly, CLL cells transduced with an adenovirus encoding CD154 (Ad-CD154) can induce CD40 activation of both transduced and nontransduced CLL cells, which then can stimulate autologous, leukemia-reactive T cells both in vitro (8) and in vivo (10–13).
Conceivably, Ad-CD154-transduced CLL cells also could induce an antibody response against CLL-associated antigens. To test for this, we examined the sera of six CLL patients before and after five biweekly infusions of autologous, Ad-CD154-transduced CLL cells.
Results
Before the infusions of autologous, Ad-CD154-transduced CLL-cells, the six patients had CLL-associated hypogammaglobulinemia with median serum levels of IgM, IgA, or IgG of 28 ± 21 mg/dl (SD), 53 ± 63 mg/dl, or 600 ± 297 mg/dl, respectively. However, 2 weeks after the final infusion, the median serum levels of IgM and IgG had increased to 77 ± 48 mg/dl (P = 0.07) and 950 ± 487 mg/dl (P < 0.05), respectively, whereas the IgA levels were not significantly changed. Five patients developed high-titer antibodies against adenovirus, with 50-, 60-, or ≈1,000-fold median increases in the titers of anti-adenovirus IgM, IgA, or IgG, respectively. The IgG response primarily was comprised of the IgG1, IgG2, and IgG3 subclasses, whereas anti-adenovirus IgG4 or IgE were not observed (data not shown).
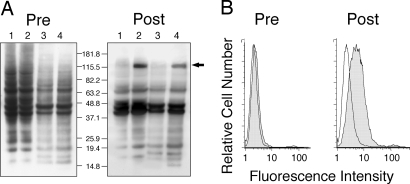
We incubated allogeneic, IgG-negative CLL cells with serial dilutions of each sera and then stained them with fluorochrome-conjugated anti-human IgG. CLL cells incubated with postinfusion sera of patients 4, 5, or 6, but not preinfusion sera or sera of healthy adults (n = 6), had increased fluorescence intensity when stained with the fluorochrome-conjugated anti-human IgG (data not shown). Although none of the preinfusion sera reacted selectively with CLL cell lysates in immunoblot analyses, the postinfusion sera that reacted with CLL cells in the flow cytometry assay reacted with a protein of ≈125 kDa in lysates of CLL cells that was not apparent in lysates of normal lymphocytes (Fig. 1A).
Fig. 1.
Evaluation for IgG Anti-CLL antibodies. (A) Lysates of membrane proteins isolated from CHO cells (lane 1), CHO-ROR1 cells (lane 2), lymphocytes of a healthy donor (lane 3), or the CLL cells of an untreated patient (lane 4) were examined by immunoblot analysis with sera obtained before (Left) or after (Right) infusions of autologous, Ad-CD154-CLL cells. The blots were treated with HRP-conjugated mouse anti-human IgG and developed with chemiluminescent substrate for autoradiography. The numbers indicate the molecular sizes of marker proteins that were electrophoresed in parallel lanes. The arrow highlights the band of ≈125 kDa identified in the lysates of CHO-ROR1 cells (lane 3) or CLL cells (lane 4) by using the postinfusion sera (Right). (B) The histograms depict the reactivity of sera with CHO cells (open histograms), or CHO-ROR1 cells (shaded histograms) before (Left) or after (Right) infusion. After washing the cells, the cell-bound antibody was detected by using a fluorochrome-conjugated goat anti-human Ig.
We evaluated the data from microarray gene-expression studies for genes used by CLL cells but not nonleukemia lymphocytes that could encode a surface protein with a molecular size of ≈125 kDa (14, 15). Among these, we focused attention on ROR1, a gene that encodes a surface, ophan-receptor type I tyrosine kinase of >100 kDa.
We examined the sequence of the ROR1 cDNA generated from the CLL cells of each of four patients. The ROR1 cDNA of one patient (A50) was identical to that of the published ROR1 cDNA (NM_005012) (16). Two other cases (A364 and A377) had ROR1 cDNA sequences that were identical to each other, but had two nucleotide differences from NM_005012 at positions 1353 and 1553. Whereas the substitution at position 1353 was conservative, the difference at position 1553 resulted in substitution of threonine for methionine at amino acid 518 of the ROR1 polypeptide sequence. This appears to represent a genetic polymorphism, as the ROR1 cDNA of these two cases matched the annotated genomic DNA contigs identified in the Human Genome Project (17). This assumption is supported by the ROR1 sequences of the fourth CLL sample (A332), which expressed equal amounts of both types of ROR1 mRNA.
The postinfusion sera of patients 4, 5, and 6, but not preinfusion sera or sera from control donors (n = 3) reacted specifically with Chinese hamster ovary (CHO) cells transduced with a ROR1-expression vector (CHO-ROR1) (Fig. 1B and data not shown). Moreover, the positive postinfusion antisera reacted with a ≈125-kDa protein in lysates of CLL cells or CHO-ROR1 cells that was not detected in lysates from normal blood lymphocytes or CHO cells (Fig. 1A). This protein was larger than the predicted molecular size of the polypeptide encoded by ROR1 (≈102 kDa), suggesting that the polypeptide expressed in CLL and CHO-ROR1 was glycosylated at deduced N-glycosylation sites.
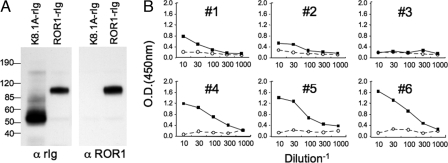
We generated a recombinant ROR1-rabbit Ig protein that had the extracellular domain of human ROR1 conjoined wth the constant region of rabbit IgG (Fig. 2A). Serial dilutions of sera obtained from patients before or after infusions of Ad-CD154-transduced CLL cells were applied to plates coated with the recombinant ROR1-rIg protein, which subsequently were developed with horseradish peroxidase (HRP) anti-human IgG. The preinfusion sera from each patient, each of 35 additional CLL patients, or 3 healthy adult donors failed to react specifically with ROR1-rIg (Fig. 2B and data not shown). However, the sera of patients that reacted with allogeneic CLL cells (patients 4, 5, and 6) each reacted with plates coated with ROR1-rIg (Fig. 2B) even when saturating amounts of rabbit Ig were added to the sera before the assay.
Fig. 2.
Anti-ROR1 antibody detected by ELISA. (A) Immunoblots of ROR1-rIg or control K8.1A-rig that subsequently were probed with goat anti-rabbit Ig (α rig) (Left) or goat anti-ROR1 Ig (α ROR1) (Right). The lanes containing the K8.1A-rig or ROR1-rig are indicated. The isolated proteins were assessed for purity by using GelCode blue stain reagent (Pierce) staining after the SDS/PAGE. The numbers indicate the molecular sizes of marker proteins run in parallel lanes of the SDS/PAGE. (B) ELISA for anti-ROR1 antibodies in sera from each patient. The open circles connected with dashed lines indicate the data with pretreatment sera, and the closed squares connected by lines indicate the data of posttreatment sera. The optical density (at 450 nm) is indicated for the serum samples at each of various dilutions (as indicated by the inverse of the dilution factor on the x axis).
To examine for surface expression of ROR1 on CLL cells, we generated anti-ROR1 antisera in mice via intradermal injection of pROR1 together with an adjuvant plasmid encoding granulocyte-monocyte-colony stimulating factor (GM-CSF) and murine CD154 as described (18). Splenocytes from mice with high-titer anti-ROR1 antisera were used to generate hybridomas with P3-X63-Ag8. One hybridoma, designated 4A5, produced mouse IgG2b mAb specific for the extracellular domain of ROR1.
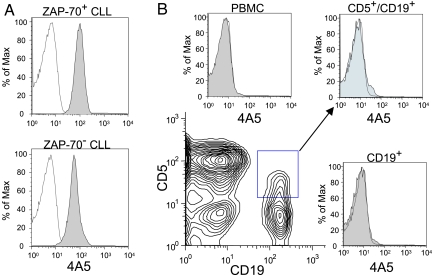
The Alexa-647-conjugated 4A5 mAb specifically stained CLL cells, but not nonleukemic leukocytes (Fig. 3). The fluorochrome-conjugated 4A5 mAb reacted with the CD5+/CD19+ CLL cells of each patient tested (n = 69), allowing for specific detection of CLL cells by flow cytometry (Fig. 3A). The 4A5 mAb reacted with ZAP-70+ CLL (n = 33) with similar intensity as with ZAP-70− CLL (n = 36). For ZAP-70+ cases the average mean fluorescence intensity ratio (MFIR) was 12.0 ± 4.4 (SD) and the median MFIR was 12.2, ranging from 2.6 to 21.2. Similarly, for ZAP-70− cases the average MFIR was 12.4 ± 5.5 (SD) and the median MFIR was 12.0, ranging from 2.4 to 25.8. In contrast, 4A5 failed to react with the lymphocytes of healthy donors, normal B cells, or CD5+/CD19+ B cells (Fig. 3B). For 4A5-stained B cells of healthy donors (n = 10) the average MFIR was 0.9 ± 0.1 (SD) and the median MFIR was 0.9, ranging from 0.8 to 1.3. The 4A5 mAb also did not react with nonleukemic blood mononuclear cells of patients with CLL, allowing for the single-color detection of CLL cells in the blood or marrow of patients with early-stage disease or minimal residual disease after therapy (data not shown).
Fig. 3.
Reactivity of 4A5 with CLL cells and nonneoplastic lymphocytes. (A) Histograms depicting the fluorescence of CLL cells stained with 4A5 (shaded histograms) or an IgG2b isotype control antibody of irrelevant specificity (open histograms). Depicted is a typical histogram for stained CLL cells that express ZAP-70 (Upper) or that lacked expression of ZAP-70 (Lower), that was detected, as described (35). (B) The peripheral blood mononuclear cells (PBMC) of a healthy adult were stained as in Fig. 4A using 4A5 or the control IgG2b along with fluorochrome-conjugated mAb specific for CD5, and CD19. The expression of CD5 (y axis) and CD19 (x axis) by the stained PBMC is depicted by using a 2D contour plot (Lower Left). The 4A5 staining of the PBMC (Upper Left), the CD5+/CD19+ B cells (Upper Right), or the total CD19+ cells (Lower Right) is provided by using gating strategies as indicated in the contour plot. The arrows lead from the electronically gated subpopulation of CD5+/CD19+B cells to the representative histogram. The shaded histograms depict the fluorescence of cells stained with 4A5 and the open histograms provide the fluorescence of cells stained with the IgG2b isotype control mAb.
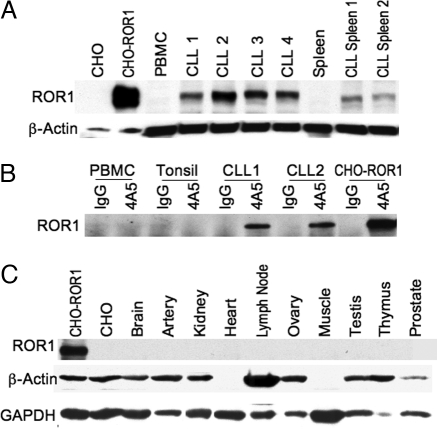
We examined lysates of normal lymphoid tissues or isolated CLL cells in immunoblot analyses by using anti-ROR1 antibodies generated against an N-terminal-region synthetic peptide. This analysis revealed a protein of ≈125 kDa in lysates of CLL cells or CHO-ROR1 cells, but not in lysates of CHO cells or mononuclear cells of healthy donors (Fig. 4A). Furthermore, 4A5 could immunoprecipitate a protein of ≈125 kDa in lysates of CLL cells or CHO-ROR1 cells (Fig. 4B). However, 4A5 could not immune precipitate this protein from lysates of blood mononuclear cells, splenocytes, or tonsillar lymphocytes of healthy adults (Fig. 4B). Moreover, we could not detect ROR1 by immunoblot analyses in the cell lysates of any other adult tissue [e.g., brain, breast, colon, heart, kidney, lung, liver, pancreas, spleen, thymus, testis, tonsil, or vascular endothelium (Fig. 4C)].
Fig. 4.
Immunoblot analyses for expression of ROR1. (A) Total cell lysates of CHO cells, CHO-ROR1 cells, CLL blood mononuclear cells (CLL samples 1–4), or CLL splenocytes (CLL spleen 1 and CLL spleen 2), blood mononuclear cells of a healthy donor (PBMC), or nonneoplastic, normal human splenocytes (Spleen) were examined by immunoblot analysis using rabbit anti-ROR1 anti-peptide antibody (Upper) or antibodies to β-actin to monitor for protein loading (Lower). The source of the tissue is indicated at the top of each lane. (B) Immunoprecipitation of ROR1 using the 4A5 mAb. Cell lysates of normal donor PBMC, normal tonsil, CLL blood mononuclear cells (CLL1 and CLL2) or CHO-ROR1 cells were incubated with the 4A5 mAb or an IgG isotype control mAb for immune precipitation using Staph protein A. The immune precipitate was evaluated by immunoblot analysis using anti-ROR1 peptide antisera. This process detected protein of ≈125 kDa in 4A5 immune precipitates prepared from CLL cell samples or CHO-ROR1 cells, but not from blood or tonsillar lymphocytes of normal donors, or the isotype control immune precipitates from any source. (C) Cells lysates were prepared from CHO-ROR1 cells, CHO cells, or human brain, artery, kidney, heart, lymph node, ovary, skeletal muscle, testis, thymus, or prostate tissue for immunoblot analyses using anti-ROR1 antibodies (Top) or antibodies specific for β-actin (Middle) or GADPH (Bottom), to monitor for protein loading. Antibodies were used to monitor for GADPH because not all tissues have detectable β-actin in this assay (e.g., cardiac and skeletal muscle).
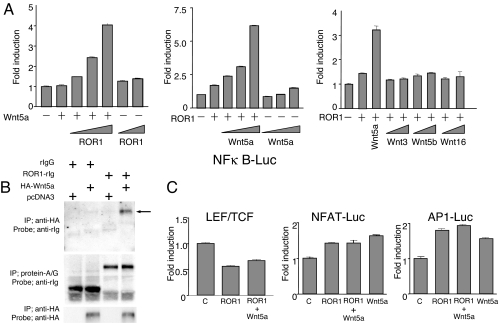
Because ROR1 shares a cysteine-rich domain with Frizzled receptors for Wnt-family proteins, we cotransduced HEK293 with reporter constructs along with vectors encoding various human Wnt factors or human ROR1. Coexpression of ROR1 in HEK293 cells with Wnt5a, but not other Wnt-family members (e.g., Wnt3, Wnt5b, or Wnt16), induced activation of NF-κB (Fig. 5A). Induction of NF-κB depended on the relative expression of ROR1 and Wnt5a, but was independent of expression of LPR5/6, which ordinarily serves as a coreceptor for Wnt receptors (19). Consistent with this finding, we found that the recombinant extracellular region of ROR1 could bind Wnt5a (Fig. 5B). However, expression of ROR1 with Wnt5a, or any of the other cotransduced Wnt factors, did not activate reporter constructs for lymphoid-enhancing-factor/T cell transcription factors (LEF/TCF), nuclear factor of activated T cells (NFAT), or activator protein-1 (AP-1) (Fig. 5C).
Fig. 5.
Functional studies on ROR1. (A) Effect of ROR1 on NF-κB activity. HEK293 cells were cotransduced with NF-κB reporter construct and β-gal vector along with expression vectors encoding ROR1, Wnt5a, Wnt3, Wnt5b, or Wnt16. (Left) Shown are the data for NF-κB activation of cells transduced without (−) or with (+) the vector encoding Wnt5a and increasing amounts of vector encoding ROR1. (Middle) Shown are the data for NF-κB activation of cells transduced without (−) or with (+) the vector encoding ROR1 and increasing amounts of vector encoding Wnt5a. (Right) Shown are the data for NF-κB activation of cells transduced without (−) or with (+) the vector encoding ROR1 and the vector encoding Wnt5a or increasing amounts of vector encoding Wnt3, Wnt5b, or Wnt16, respectively. Each bar provides the mean fold induction over baseline of triplicate test samples ± SE. (B) In vitro binding of ROR1 to Wnt5a. Cells were transduced with vectors encoding an HA-tagged Wnt5a recombinant protein (designated HA-Wnt5a). The conditioned media of such cells were incubated with or without ROR1-rIg or rIgG before immune precipitation using anti-HA antibodies (Top and Bottom) or protein-A/G (Middle). The blots were probed with antibodies specific for rIgG (Top and Middle) or anti-HA (Bottom). At the top of each lane is the combination of reagents used in the immune precipitation studies. + indicates that the constituent designated on the left was used in preparing the sample for that specific lane. The lanes marked with + for pcDNA3 used conditioned supernatant from cells transduced with control vector that did not encode Wnt5a or ROR1. (C) Effect of ROR1 on lymphoid enhancer-binding factor/T cell factor (LEF/TCF)-sensitive luciferase reporter gene TOPFLASH (Left), nuclear factor of activated T cells (NF-AT) (Center), or activator protein 1 (AP-1) (Right). HEK293 cells were cotransduced with indicated reporter constructs, a vector encoding β-gal (control labeled C), or vectors encoding ROR1 and/or Wnt5a. Each bar provides the mean fold induction over baseline of triplicate test samples ± SE.
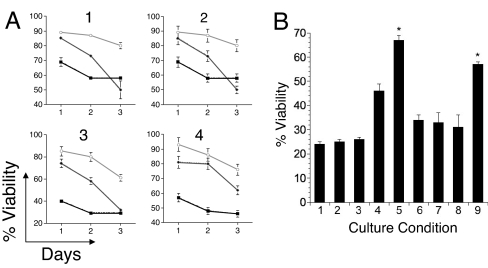
CLL cells lack expression of Wnt5a (19). As such, we examined the effect of Wnt5a on CLL cells in vitro. CLL cells were cultured alone or together with CHO cells or CHO-Wnt5a cells, and the viability of the CLL cells was monitored over time. The viability of CLL cells cocultured with CHO cells or CHO-Wnt5a cells was significantly greater than that of CLL cells cultured alone, particularly after 1 day in culture. However, in each case, the viability of CLL cells cocultured with CHO-Wnt5a cells was significantly greater than that of CLL cells cocultured with CHO cells, particularly at later time points (Fig. 6A).
Fig. 6.
Influence of Wnt5a on CLL cell viability. (A) Effect of Wnt5a on the viability of CLL cells cultured in vitro. CLL cells from each of four unrelated patients, designated CLL 1, CLL 2, CLL 3, and CLL 4, were cultured alone (■) or together with CHO cells (♦), or CHO-Wnt5a cells (○). The percent viability of the CD19+ CLL cells, indicated on the ordinate, was assessed by flow cytometry on days 1, 2, and 3 of culture. Each data point represents the mean value of quadruplicate samples cultured in parallel ± SE. (B) CLL cells were cultured for 2 days in RPMI medium 1640 containing 20% human serum, either alone or together with CHO cells or CHO-Wnt5a cells and then assessed for viability by flow cytometry. The bars indicate the mean percent viability of the CD19+ CLL cells of quadrulicate wells for each culture condition. Except for condition 1, all cultures had serum samples from patient 4 collected either before or 2 weeks after the last infusion of autologous Ad-CD154-trasduced CLL cells. CLL cells were cultured alone in media containing normal human serum (condition 1), preinfusion serum (condition 2), or postinfusion serum (condition 3). For culture conditions 4, 6, and 8 the CLL cells were cocultured with CHO cells and for culture conditions 5, 7, and 9 the CLL cells were cocultured with CHO-Wnt5a cells. Cultures 4 and 5 used preinfusion serum, cultures 6 and 7 used postinfusion serum that had been absorbed with CHO cells, and cultures 8 and 9 used postinfusion serum that had been absorbed with CHO-ROR1 cells to remove anti-ROR1 binding activity. The error bars depict the SE about the mean of quadruplicate wells cultured in parallel. The asterisks indicate that the viability of CLL cells in those culture condition was significantly greater than that of the other culture conditions by Bonferroni t test (P < 0.05).
Neither 4A5 nor the preinfusion or postinfusion sera of any of the patients enhanced or reduced the viability of CLL cells relative to that of CLL cell cultured in media with normal human serum or FBS (Fig. 6B; culture conditions 1, 2, or 3). We examined whether the postinfusion anti-ROR1 antisera affected the survival of CLL cells cocultured with CHO cells or CHO-Wnt5a cells. Although the preinfusion sera of each patient did not affect CLL-cell survival, the postinfusion sera of patients 4 and 5 neutralized the capacity of CHO-Wnt5a cells to enhance the viability of CLL cells over that afforded by culture with CHO cells lacking Wnt5a. For example, CLL cells cocultured with CHO-Wnt5a had significantly higher viability than CLL cells cocultured with CHO cells or CLL cells cultured alone in media containing 20% preinfusion serum (Fig. 6B; culture condition 5 relative to conditions 4 or 2). However, the postinfusion serum could neutralize the capacity of CHO-Wnt5a to promote CLL cell survival relative to that of CHO cells or media alone, even when the postinfusion serum was absorbed with CHO cells before use in the assay (Fig. 6B; culture condition 7 relative to conditions 6 or 3). On the other hand, when we used media with postinfusion serum that had been absorbed with CHO-ROR1 cells, we found that the CHO-Wnt5a cells again provided a significant survival advantage to the CLL cells relative to that provided by CHO cells or media alone (Fig. 6B; culture condition 9 relative to conditions 8 or 3).
Discussion
Most patients infused with autologous Ad-CD154-transduced CLL cells developed antibody responses against adenovirus and some developed anti-CLL autoantibodies, which we found reacted with ROR1. Although ROR1 was expressed on all CLL cases examined, this surface protein was not found on normal adult tissues or lymphoid cells, including CD5+ B cells. As such, ROR1 represents a CLL-associated antigen against which patients can develop antibodies.
Sequence analyses revealed no apparent somatic mutations within the ROR1 sequence in CLL. Moreover, we found CLL cells could coexpress either of two ROR1 alleles identified earlier (16, 17), indicating that the disease is not associated with one allele. This finding also makes it unlikely that expression of ROR1 in CLL is secondary to mutations in the promoter/enhancer region, translocations, and/or mutations that affect transcript stability or processing. Nevertheless, the restricted expression of ROR1 in CLL suggests it plays a role in pathogenesis.
ROR1 initially was identified by using oligonucleotide primers targeting sequences encoding tyrosine kinase domains of different proteins (20). This protein appears highly conserved throughout evolution (21, 22). In rodents, ROR1 is expressed primarily in developing cephalic neural crest in the dorsal part of the diencephalons and mid-hind brain boundary during embryogenesis (23). In most species examined, ROR1 expression attenuates after embryonic development, becoming negligible at term (20, 24).
Work in Caenorhabditis elegans indicated that the ROR1-type kinases might be involved in the regulation of cell motility and in asymmetric cell division during embryogenesis (25). Furthermore, the ROR protein in C. elegans, CAM-1/KIN-8, apparently has both kinase-dependent and kinase-independent functions (25, 26). ROR-family receptor tyrosine kinases are characterized by the intracellular tyrosine kinase domains, highly related to those of the Trk-family receptor tyrosine kinases, and by the extracellular Frizzled-like cysteine-rich domains and kringle domains, which are common to receptors of the Wnt family members. An ortholog to ROR1, namely ROR2, was found to interact with Wnt5a to activate noncanonical Wnt signaling (27, 28).
We found that ROR1 interacts physically with Wnt5a independent of LPR5/6, which could induce activation of NF-κB in a mutually dose-dependent fashion (Fig. 5A). This activity also was independent of expression of LPR5/6, which ordinarily serves as a coreceptor for Wnt receptors (19).
CLL cells derive a survival benefit from interactions with marrow stroma (29), nurse-like cells (30), or dendritic cells (31), which are found in the lymphoid tissues of patients with this disease. Conceivably ROR1 could play a role in such interactions. It is noteworthy that dendritic cells also express high levels of Wnt5a (32). Although there are other receptors for Wnt5a, we found that anti-ROR1 antisera specifically could neutralize the survival advantage afforded by coculture with CHO-Wnt5a cells (Fig. 6B). As such, ROR1 might enhance the capacity of CLL cells to receive survival signals from its microenvrionment and provide a novel target for treatment of this disease.
Materials and Methods
Patients.
Six patients with advanced-stage CLL each received five biweekly i.v. infusions of 3–6 × 108 autologous CLL cells transduced ex vivo with a replication-defective, serotype-5 adenovirus encoding murine CD154 as described (11). Sera were collected before the initial infusion and at 2 weeks after the final infusion.
Isolation and Sequencing of ROR1 cDNA.
Total cellular RNA was isolated with RNeasy reagents (Qiagen). First-strand cDNA was synthesized by using an oligo(dT) primer and SuperScript II RT (Invitrogen). After treatment with RNase H the cDNA was purified by using QIAquik columns (Qiagen) and used for PCR amplification of ROR1 with 200 μM dNTPs, 3% DMSO, Phusion Hot Start DNA polymerase (New England Biolabs), 1× Phusion GC Buffer (New England Biolabs), and 250 μM each of oligonucleotide primers ROR1-F (5′-CGAGAGGAGGAATGCAC-3′) and ROR1-R (5′-ATACCACATTTACAAAAGTTGTG-3′). PCR cycling parameters were 98°C for 2 min, followed by 35 cycles of 98°C for 15 s, 59°C for 30 s, and 72°C for 1 min. The PCR products were size-selected in 0.8% agarose/0.5 μg/ml of ethidium bromide (Invitrogen) and purified by using QIAquik columns. PCR products were sequenced directly by using the fluorescence-dideoxy-chain-termination method and an automated nucleic acid sequence analyzer (Applied Biosystems). Nucleotide sequences were analyzed by using DNASTAR and compared with data deposited in the GenBank sequence databases by using BLAST (www.ncbi.nlm.nih.gov/BLAST) or the cDNA reference sequence (NM_005012).
Plasmids and DNA Immunization.
We inserted the human ROR1 cDNA (Origene Technologies) into pcDNA3 (Invitrogen) to generate pROR1. We cloned the 5′ portion of the ROR1 cDNA encoding the ROR1 extracellular domain into the pcDNA3-zeocin vector (Invitrogen), placing it upstream and in-frame with the cDNA encoding the rabbit IgG Fc region (rIg) to generate pROR1-rIgG. The plasmids pCD40L or pGM-CSF were as described (18, 33). We inserted the cDNA encoding human Wnt5a into pUSEamp (Upstate Biotechnology) to generate pWnt5a.
Plasmid DNA was purified by using the Qiagen EndoFree Plasmid Mega Kit with inclusion of a 10% Triton X-114 (Sigma) extraction step before application of the bacterial lysate onto the column. Six-week-old female BALB/c mice received three biweekly intradermal injections of plasmid DNA in 50 μl of isotonic saline containing 100 μg of pROR1 and 50 μg each of pGM-CSF pCD40L near the base of the tail with a 28G-insulin syringe (Becton Dickinson) as described (18). Blood samples were collected via retro-orbital puncture before the first injection and 1 week after the last injection and stored at −20°C before analysis.
Cells and Cell Lines and Recombinant Proteins.
Blood samples were collected after written, informed consent, as per the Declaration of Helsinki. Mononuclear cells were isolated by using Ficoll-Hypaque (Amersham Pharmacia). Cells were suspended in FCS containing 5% DMSO for storage in liquid nitrogen until use. Upon thawing the viability of the mononuclear cells was ≥85%, as assessed by exclusion of propidium iodide (PI) (Invitrogen).
CHO cells were from the American Type Tissue Culture Collection. To generate CHO-ROR1, CHO-ROR1-rIgG, or CHO-Wnt5a cells, the CHO cells were transduced with pROR1, pROR1-rIgG, or pWnt5a, respectively, by using Lipofectamine 2000 (Invitrogen). Cells transduced with pROR1 or pROR1-rIgG were cultured in DMEM (Gibco) supplemented with 10% FCS for 24 h and then placed in selection media containing G418 (250 μg/ml) or Zeocin 300 μg/ml) (Invitrogen). Cloned cells were evaluated for expression of ROR1 or ROR1-rIgG by immunoblot analysis. The cells were adapted to suspension culture in IMGX II medium (HyClone). Suspended CHO-ROR1-rIg cells were cultured in ProCHO-5 medium (Cambrex Bio Science), and recombinant ROR1-rIg was purified from the culture supernatant by using protein A Sepharose (Pierce Biotechnology). The purity of the isolated protein was assessed by PAGE and immunoblot analysis. The K8.1 glycoprotein of Kaposi sarcoma-associated herpes virus protein fused with the rIg Fc recombinant protein (K8.1A-rIg) was used as a control recombinant rIg protein. The splenocytes of mice that generated anti-ROR1 antibodies were fused with P3-X63-Ag8 to generate hybridomas that were selected as described (34). One hybridoma, designated 4A5, produced IgG2b anti-ROR1 mAb.
Immunoblot Analyses.
Total cell lysates were made by incubation cells in a RIP lysis buffer containing 1% Triton X-100, 50 mM Tris·HCl (pH 7.5), 100 mM NaCl, 50 mM NaF, 5 mM EDTA with protease inhibitors (10 μg/ml aprotinin, 10 μg/ml leupeptin, 10 μg/ml pepstatin, and 1 mM phenylmethylsulfonyl fluoride), and phosphatase inhibitors (40 mM glycerophosphate and 1 mM sodium orthovanadate) (Roche). Cell lysates were separated 7.5% or 5–15% gradient SDS/PAGE and transferred onto Immobilon-P membranes (Millipore). Nonspecific binding sites were blocked with 10% powdered milk before treating the membranes with patient sera (diluted 1:10 in PBS containing 5% FBS) or with rabbit (Cell Signaling Technology) or goat (R&D Systems) anti-ROR1-peptide antibodies in 5% FBS for overnight incubation at 4°C. Antibodies to human Wnt5a were obtained from Cell Signaling Technologies. For detection of membrane-bound human, rabbit, or goat IgG, the washed membranes, respectively, were incubated with mouse anti-human IgG, anti-rabbit Ig, or anti-goat Ig that was conjugated to HRP (Santa Cruz Biotechnology) for subsequent development with Super Signal West Femto Chemiluminescent Substrate (Pierce) for autoradiography with Super RX film (Fuji).
Flow Cytometry.
To analyze sera for IgG anti-CLL antibodies CLL cells were stained with serial dilutions of antisera for 30 min at 4°C in staining media (SM; RPMI-1640/0.5% BSA). The cells were washed twice in SM and then counterstained with phycoerythrin (PE) or allophycocyanin (APC)-labeled mouse anti-human IgG, fluorescein-conjugated anti-CD3, and/or PE or APC-conjugated anti-CD19 or anti-CD5 for 30 min at 4°C. In others studies, CLL cells, CHO cells, and/or CHO-ROR1 cells were each stained with sera collected from treated patients or healthy adult donors, or antisera generated in mice that had been immunized against ROR1 via DNA immunization. Cells were analyzed by using a FACSCalibur (Becton Dickinson) with FlowJo software (Tree Star).
We conjugated 4A5 and an IgG2b isotype control mAb of irrelevant specificity with Alexa Fluor 647 (Invitrogen). The MFIR is the mean fluorescence intensity of cells stained with specific antibody divided by the mean fluorescence intensity of the same cell population stained with fluorochrome-conjugated isotype control mAb. Intracellular staining for ZAP-70 was as described (35).
ELISA and Ig Measurement.
Polystyrene microtiter plates were incubated overnight at 4°C with ROR1-rIg (at 5 μg/ml PBS, pH 7.4), rIg (at 5 μg/ml PBS), or adenovirus that previously had been heated to 65°C for 15 min (at 108 virus particles per ml of PBS), to measure for antibodies reactive with ROR1-rIg, rIg, or adenovirus, respectively. After washing the plates three times with 0.05% Tween-20 in PBS and treatment with 2% BSA/PBS for 1 h at room temperature, serial dilutions of antisera in 2% BSA/PBS were added to individual wells and incubated for 1 h at room temperature or overnight at 4°C. After washing the plates three times with 2% BSA/PBS, HRP or alkaline phosphatase (AP)-conjugated goat antibody specific for either human Ig, IgG, IgA, IgM, IgG1, IgG2, IgG3, or IgG4 (Southern Biotechnology) were added to separate wells. SureBlue TMB Microwell Substrate (KPL) or SIGMA FAST pNPP (Sigma) were added to the wells of washed plates to develop antibodies labeled with HRP or AP, respectively. Replicate wells of plates were analyzed by using a SpectraMax250 plate reader (Molecular Devices). Total serum IgM, IgA, and IgG levels was measured by using automated nephelometric and immunoturbidimetric assays as described (36).
Reporter Assay.
Reporter assays were performed as described (19). Briefly, HEK293 cells were transduced in 12-well plates by using FuGENE (Roche), and 0.5 μg of reporter plasmid, 0.1–0.2 μg of the control plasmid pCMX β-gal, 100–200 ng of the various expression plasmids, and carrier DNA pBluescriptKSII, for a total of 1 μg per well. The luciferase values were normalized for variations in transduction efficiency by using the β-gal internal control and are expressed as fold stimulation of luciferase activity, compared with the designated control cultures. Results are representative of a minimum of three independent experiments.
Cell Culture Assays.
CHO cells or CHO-Wnt5a cells were cultured in DMEM supplemented with 10% FBS. These cells were plated into separate wells of 24-well culture plates in 500 μl of media at 5 × 105 cells per well. Viably frozen primary CLL cells from different patients were washed and then suspended in RPMI medium 1640 at 1 × 107 cells/ml. The viability of each CLL cell population exceeded 85%. CLL cells at 5 × 106 cells per well were seeded onto wells containing media alone or CHO cells or CHO-Wnt5a cells. The viability of the CD19+ CLL cells in each of quadruplicate wells for each condition and time point was examined by flow cytometry PI and DiOC6 as described (37).
Patient sera collected before or 2 weeks after staining the cells with postinfusions of autologous Ad-CD154-transduced CLL cells were examined for their capacity to alter the viability of primary CLL cells cultured alone or together with CHO cells or CHO-Wnt5a cells. For studies using absorbed serum samples, 20% human serum in RPMI medium 1640 was used to suspend cell pellets of CHO cells or CHO-ROR1 cells to a final cell concentration of 2 × 106/ml. The samples were incubated for 30 min at 4°C and then spun at 200 × g for 4 min. The supernatant from each was removed and used as culture media. For this process 2.5 × 106 CLL cells in 250 μl of media were added to each well of a 48-well culture plate that contained 250 μl of culture media alone or 250 μl of culture media with 2.5 × 105 CHO cells or CHO-Wnt5a cells. Quadruplicate wells for each condition were harvested after 48 h, and the viability of the cultured CD19+ CLL cells was measured by flow cytometry after staining the cells with PI and DiOC6 as described (37).
ACKNOWLEDGMENTS.
We thank Esther Avery, Dennis Young, Andrew Greaves, and Robier Aguillon for assistance in this work and Dr. Yu-Tsueng Liu (University of California at San Diego) for the K8.1ArIg vector. This work was supported in part by National Institutes of Health Grant PO1-CA081534 and a Specialized Centers of Research grant (to T.J.K.) from the Leukemia and Lymphoma Society of America.
Footnotes
The authors declare no conflict of interest.
References
- 1.Sinisalo M, et al. Response to vaccination against different types of antigens in patients with chronic lymphocytic leukemia. Br J Haematol. 2001;114:107–110. doi: 10.1046/j.1365-2141.2001.02882.x. [DOI] [PubMed] [Google Scholar]
- 2.van der Velden AM, et al. Influenza virus vaccination and booster in B-cell chronic lymphocytic leukaemia patients. Eur J Intern Med. 2001;12:420–424. doi: 10.1016/s0953-6205(01)00149-2. [DOI] [PubMed] [Google Scholar]
- 3.Hartkamp A, Mulder AH, Rijkers GT, van Velzen-Blad H, Biesma DH. Antibody responses to pneumococcal and hemophilus vaccinations in patients with B-cell chronic lymphocytic leukemia. Vaccine. 2001;19:1671–1677. doi: 10.1016/s0264-410x(00)00409-6. [DOI] [PubMed] [Google Scholar]
- 4.Lotz M, Ranheim E, Kipps TJ. Transforming growth factor β as endogenous growth inhibitor of chronic lymphocytic leukemia B cells. J Exp Med. 1994;179:999–1004. doi: 10.1084/jem.179.3.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ranheim EA, Cantwell MJ, Kipps TJ. Expression of CD27 and its ligand, CD70, on chronic lymphocytic leukemia B cells. Blood. 1995;85:3556–3565. [PubMed] [Google Scholar]
- 6.Cantwell M, Hua T, Pappas J, Kipps TJ. Acquired CD40-ligand deficiency in chronic lymphocytic leukemia. Nat Med. 1997;3:984–989. doi: 10.1038/nm0997-984. [DOI] [PubMed] [Google Scholar]
- 7.Ranheim EA, Kipps TJ. Activated T cells induce expression of B7/BB1 on normal or leukemic B cells through a CD40-dependent signal. J Exp Med. 1993;177:925–935. doi: 10.1084/jem.177.4.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kato K, Cantwell MJ, Sharma S, Kipps TJ. Gene transfer of CD40 ligand induces autologous immune recognition of chronic lymphocytic leukemia B cells. J Clin Invest. 1998;101:1133–1141. doi: 10.1172/JCI1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gorgun G, Holderried TA, Zahrieh D, Neuberg D, Gribben JG. Chronic lymphocytic leukemia cells induce changes in gene expression of CD4 and CD8 T cells. J Clin Invest. 2005;115:1797–1805. doi: 10.1172/JCI24176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kater AP, van Oers MH, Kipps TJ. Cellular immune therapy for chronic lymphocytic leukemia. Blood. 2007;110:2811–2818. doi: 10.1182/blood-2007-01-068932. [DOI] [PubMed] [Google Scholar]
- 11.Wierda WG, et al. CD40-ligand (CD154) gene therapy for chronic lymphocytic leukemia. Blood. 2000;96:2917–2924. [PubMed] [Google Scholar]
- 12.Wierda WG, Kipps TJ. Gene therapy and active immune therapy of hematologic malignancies. Best Pract Res Clin Haematol. 2007;20:557–568. doi: 10.1016/j.beha.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 13.Messmer D, Kipps TJ. CD154 gene therapy for human B-cell malignancies. Ann NY Acad Sci. 2005;1062:51–60. doi: 10.1196/annals.1358.008. [DOI] [PubMed] [Google Scholar]
- 14.Klein U, et al. Gene expression profiling of B cell chronic lymphocytic leukemia reveals a homogeneous phenotype related to memory B cells. J Exp Med. 2001;194:1625–1638. doi: 10.1084/jem.194.11.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rosenwald A, et al. Relation of gene expression phenotype to immunoglobulin mutation genotype in B cell chronic lymphocytic leukemia. J Exp Med. 2001;194:1639–1647. doi: 10.1084/jem.194.11.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paganoni S, Ferreira A. Neurite extension in central neurons: A novel role for the receptor tyrosine kinases Ror1 and Ror2. J Cell Sci. 2005;118:433–446. doi: 10.1242/jcs.01622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- 18.Burger JA, Mendoza RB, Kipps TJ. Plasmids encoding granulocyte-macrophage colony-stimulating factor and CD154 enhance the immune response to genetic vaccines. Vaccine. 2001;19:2181–2189. doi: 10.1016/s0264-410x(00)00382-0. [DOI] [PubMed] [Google Scholar]
- 19.Lu D, et al. Activation of the Wnt signaling pathway in chronic lymphocytic leukemia. Proc Natl Acad Sci USA. 2004;101:3118–3123. doi: 10.1073/pnas.0308648100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Masiakowski P, Carroll RD. A novel family of cell surface receptors with tyrosine kinase-like domain. J Biol Chem. 1992;267:26181–26190. [PubMed] [Google Scholar]
- 21.Wilson C, Goberdhan DC, Steller H. Dror, a potential neurotrophic receptor gene, encodes a Drosophila homolog of the vertebrate Ror family of Trk-related receptor tyrosine kinases. Proc Natl Acad Sci USA. 1993;90:7109–7113. doi: 10.1073/pnas.90.15.7109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Katoh M. Comparative genomics on ROR1 and ROR2 orthologs. Oncol Rep. 2005;14:1381–1384. doi: 10.3892/or.14.5.1381. [DOI] [PubMed] [Google Scholar]
- 23.Matsuda T, et al. Expression of the receptor tyrosine kinase genes, Ror1 and Ror2, during mouse development. Mech Dev. 2001;105:153–156. doi: 10.1016/s0925-4773(01)00383-5. [DOI] [PubMed] [Google Scholar]
- 24.Paganoni S, Ferreira A. Expression and subcellular localization of Ror tyrosine kinase receptors are developmentally regulated in cultured hippocampal neurons. J Neurosci Res. 2003;73:429–440. doi: 10.1002/jnr.10674. [DOI] [PubMed] [Google Scholar]
- 25.Forrester WC, Dell M, Perens E, Garriga G. A C. elegans Ror receptor tyrosine kinase regulates cell motility and asymmetric cell division. Nature. 1999;400:881–885. doi: 10.1038/23722. [DOI] [PubMed] [Google Scholar]
- 26.Kim C, Forrester WC. Functional analysis of the domains of the C. elegans Ror receptor tyrosine kinase CAM-1. Dev Biol. 2003;264:376–390. doi: 10.1016/j.ydbio.2003.09.007. [DOI] [PubMed] [Google Scholar]
- 27.Oishi I, et al. The receptor tyrosine kinase Ror2 is involved in noncanonical Wnt5a/JNK signalling pathway. Genes Cells. 2003;8:645–654. doi: 10.1046/j.1365-2443.2003.00662.x. [DOI] [PubMed] [Google Scholar]
- 28.Mikels AJ, Nusse R. Purified Wnt5a protein activates or inhibits β-catenin-TCF signaling depending on receptor context. PLoS Biol. 2006;4:e115. doi: 10.1371/journal.pbio.0040115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lagneaux L, Delforge A, De Bruyn C, Bernier M, Bron D. Adhesion to bone marrow stroma inhibits apoptosis of chronic lymphocytic leukemia cells. Leuk Lymphoma. 1999;35:445–453. doi: 10.1080/10428199909169609. [DOI] [PubMed] [Google Scholar]
- 30.Burger JA, et al. Blood-derived nurse-like cells protect chronic lymphocytic leukemia B cells from spontaneous apoptosis through stromal cell-derived factor-1. Blood. 2000;96:2655–2663. [PubMed] [Google Scholar]
- 31.Pedersen IM, et al. Protection of CLL B cells by a follicular dendritic cell line is dependent on induction of Mcl-1. Blood. 2002;100:1795–1801. [PubMed] [Google Scholar]
- 32.Lehtonen A, et al. Gene expression profiling during differentiation of human monocytes to macrophages or dendritic cells. J Leukoc Biol. 2007;82:710–720. doi: 10.1189/jlb.0307194. [DOI] [PubMed] [Google Scholar]
- 33.Mendoza RB, Cantwell MJ, Kipps TJ. Immunostimulatory effects of a plasmid expressing CD40 ligand (CD154) on gene immunization. J Immunol. 1997;159:5777–5781. [PubMed] [Google Scholar]
- 34.Kipps TJ, Herzenberg LA. In: Handbook of Experimental Immunology. Herzenberg LA, Blackwell C, Herzenberg LA, editors. Oxford: Blackwell; 1986. pp. 108.101–108.109. [Google Scholar]
- 35.Rassenti LZ, et al. ZAP-70 compared with immunoglobulin heavy-chain gene mutation status as a predictor of disease progression in chronic lymphocytic leukemia. N Engl J Med. 2004;351:893–901. doi: 10.1056/NEJMoa040857. [DOI] [PubMed] [Google Scholar]
- 36.Warren JS. In: Manual of Molecular and Clinical Laboratory Immunology. Detrick B, Hamilton RG, Folds JD, editors. Washington DC: Am Soc Microbiol; 2006. pp. 69–74. [Google Scholar]
- 37.Hu D, Kipps TJ. Reduction in mitochondrial membrane potential is an early event in Fas-independent CTL-mediated apoptosis. Cell Immunol. 1999;195:43–52. doi: 10.1006/cimm.1999.1513. [DOI] [PubMed] [Google Scholar]