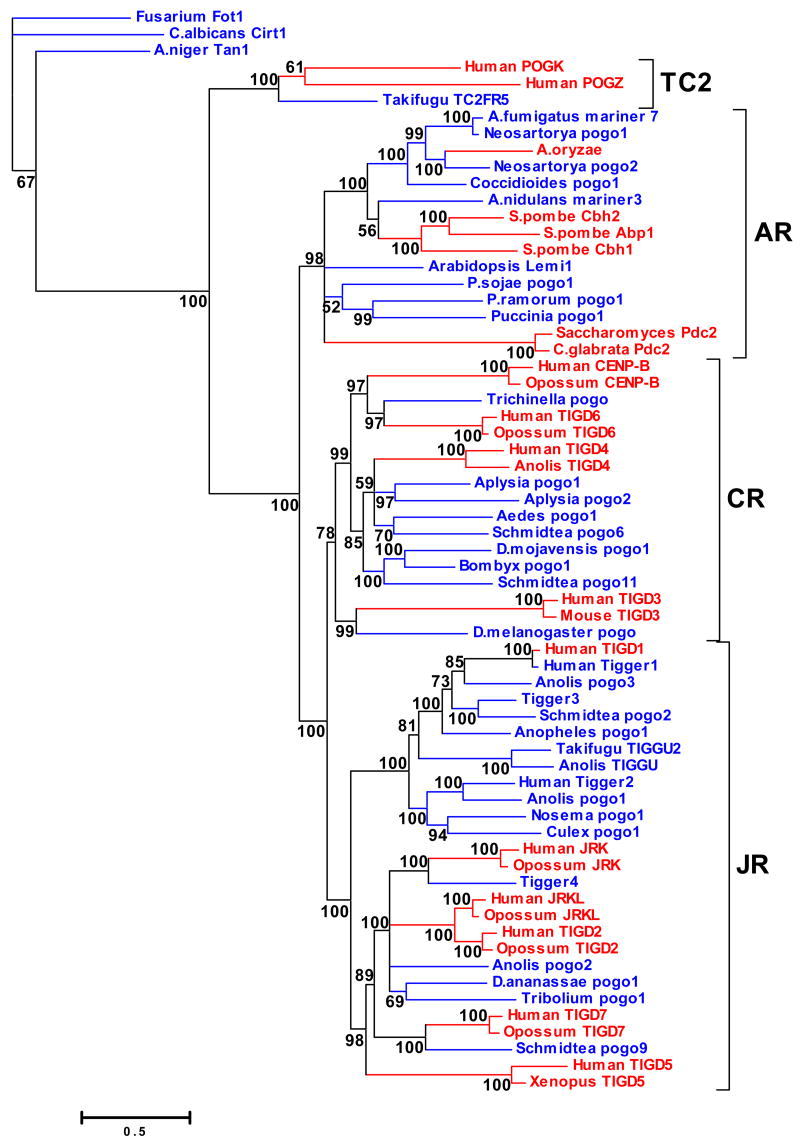
Figure 2.
Phylogenetic tree of pogo-like transposases and pogo-derived genes. Protein sequences were aligned using the MAFFT package and the alignment refined manually with Bioedit to produce a final multialignment of about 310 residues. The tree shown here was inferred using the Bayesian algorithms implemented in MrBayes as described in Methods. Numbers in the nodes show posterior probabilities. pogo-derived genes and pogo-like transposons are highlighted in red and blue, respectively. Detailed information about each sequence are reported in table 1 and table 2. CR: CENP-B-related clade; AR:Abp1-related clade; JR: JERKY-related clade: TC2: TC2-related clade. Fot1, Cirt1 and Tan1 belong to the Fot1 clade.