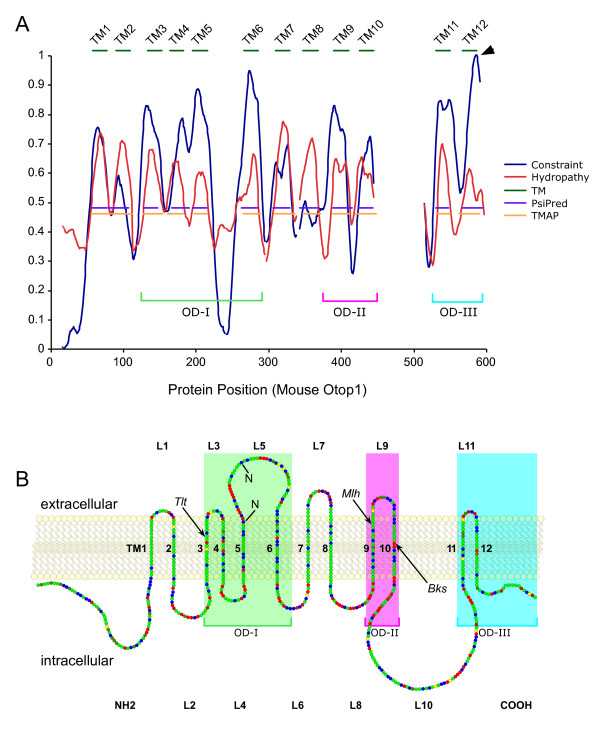
Figure 2.
Predicted secondary structure and topologic model for Otop1 insertion into the lipid bilayer. A) Hydrophobicity (red) and evolutionary constraint (blue) are plotted against the amino-acid position of mouse Otop1. A total of 12 evolutionarily constrained regions are found in the ODP family that are highly hydrophobic and have a helical structure consistent with TM domains (dark green), as predicted by TMAP (orange) and PsiPred (purple). Green, pink, and blue brackets define the highly conserved subdomains: Otopetrin Domain-I, -II, and -III (OD-I, OD-II, and OD-III, respectively). B) Linear model of mouse Otop1a inserted in a lipid bilayer, in which each amino acid is represented as a circle and the chemical properties of amino-acids are denoted by color: charged residues (red), polar residues (blue), and non-polar residue (green). Cysteine (yellow) and proline (dark green) are noted. The two consensus N-glycosylation sites (N) are indicated in loop 5. The predicted intracellular and extracellular loops and TM domains are numbered L1 to L11 and TM1 to TM12, respectively. The locations of the tlt, mlh, and bks mutations are noted by arrows. The three OD subdomains are shaded with the color code used in A.