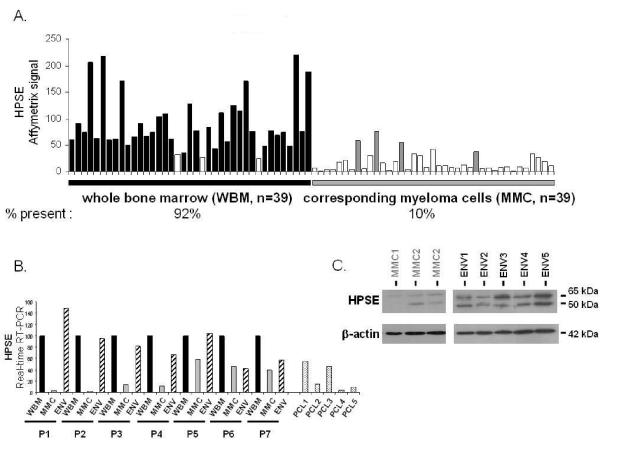
Figure 4. Comparison of HPSE expression between WBM and the corresponding purified MMC of patients.
A. Expression of HPSE was determined in the WBM of 39 myeloma patients as well as in the myeloma cells purified from the bone marrow of the same patients, using Affymetrix U1332 Plus 2.0 microarrays. White histograms indicate that the Affymetrix call is “absent”; black (WBM) and grey (MMC) histograms indicate that it is “present”. B. HPSE expression was measured by real-time RT-PCR in the whole bone marrow of seven MM patients (WBM, black bars), in the corresponding MMC (grey bars), in the corresponding microenvironment depleted from myeloma cells (<2% MMC, ENV, crosshatched bars) and in MMC from five patients with PCL. HPSE expression was normalized to that of GAPDH. For each patient, the WBM sample was used as a reference and was assigned the arbitrary value of 100. For the five patients with PCL, the WBM sample of patient 1 was used as a reference. The median percentage of plasma cells in bone marrow aspirates from the seven patients with intramedullary myeloma was 8,5% (range=6–40). C. HPSE protein expression was determined in microenvironment cells depleted from MMC (< 2% MMC, ENV1-5) of five patients and in purified MMC of three other patients (MMC1-3). Cells were lysed and the lysates were separated on a 12% SDS-PAGE and analyzed by Western blot with a polyclonal anti-HPSE antibody. Both the 65 kDa MW pro-enzyme and the 50 kDa MW active form were identified. β-actin was used as a loading control. Results are from two separate western blots, one with 3 purified MMC, the other one with the 5 microenvironment cells. The HPSEpos XG-2 HMCL was used as a positive HPSE protein control in the two blots (results not shown). kDa, molecular weight in thousands.