Fig. 3.
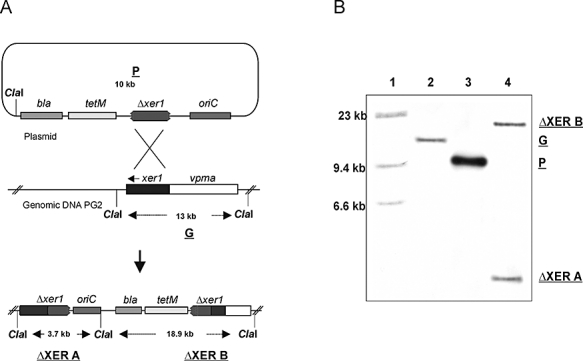
Disruption of xer1 recombinase in M. agalactiae. A. Schematic representation of the integration of disruption plasmid pR3 (P) into the genomic DNA of type strain PG2 to generate the PLMU and PLMY clones. A 13 kb ClaI fragment of PG2 (G) consists of the known 9.6 kb vpma locus including the six vpma genes (white region) and the complete xer1 gene (black shaded region). A single putative homologous recombination event between the partial xer1 sequence carried by the plasmid pR3 and the chromosomal xer1 region is represented by crossed lines. This crossing over would lead to the integration of pR3 into the chromosome and would segregate the xer1 region onto two ClaI fragments ΔXERA and ΔXERB. B. Southern blot hybridization showing the localization of pR3 at the chromosomal xer1 locus of M. agalactiae PG2. ClaI-digested DNA of xer1 disruptant (PLMU or PLMY) (lane 4), disruption plasmid pR3 (lane 3) and wt strain PG2 (lane 2) were probed with a xer1-specific DIG-labelled fragment. λ-HindIII DNA size marker (lane 1).
