Fig. 5.
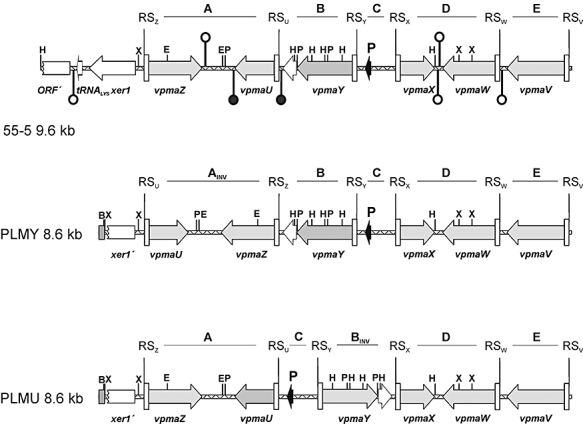
Arrangement of the ORFs, terminator structures and inversions in the vpma loci of M. agalactiae type strain PG2 clonal variant 55-5, PLMY and PLMU. Grey arrows represent the vpma genes, whereby a dark grey arrow denotes the vpma gene which is transcribed in the respective clonal variant or PLM. The black arrows indicate the location of the unique promoter (P) in each vpma locus. Putative recombination sites are indicated by white rectangles (RS) and denote sequences from −1 to −72 relative to the start codons of the corresponding vpma genes. White arrows indicate ORFs other than vpma genes, like the intact xer1 recombinase (xer1) and a tRNA (tRNALYS) gene in 55-5, whereas the discontinued white rectangles represent the promoter proximal regions of the disrupted xer1 genes (xer′) in the PLMs and a partial ORF (ORF′) in 55-5. A small grey rectangle indicates sequences derived from the integrated plasmid pR3. Non-translated repeats of the vpmaY genes are indicated with a dotted-lined white arrow. Terminator structures are indicated with symbolized hairpins, whereby Rho-independent terminator structures, for which experimental evidence is given, are drawn with a black-filled loop and are white-filled otherwise. Sequence-identical blocks between PLMs and 55-5 are designated A to E and are indicated with INV when inverted compared with 55-5. B, BamHI; E, EcoRI; H, HindIII; P, PstI; X, XbaI.
