Figure 4. Molecular determinants of Cav2.1 channel inactivation.
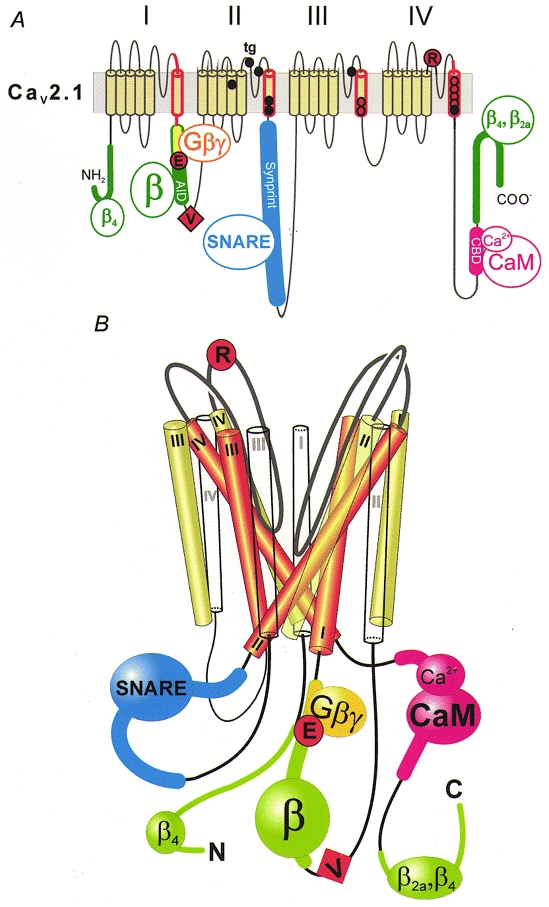
A, inactivation of Cav2.1 channels is affected by mutations in the α12.1-subunit and proteins interacting with intracellular loops. The scheme highlights crucial segments and the location of some key point mutations: segment IS6 and adjacent intra- and extracellular sequences (red; Zhang et al. 1994); mutations in the putative pore-lining region of segments IIIS6 and IVS6 (red circles; Hering et al. 1996, 1998; Sokolov et al. 2000); point mutations associated with FHM, shown as black circles (see Kraus et al. 1998, 2000, for functional studies); point mutations in the intracellular I-II loop (G-protein-interaction motif (yellow): QQIER→QQIEE; Herlitze et al. 1997) and the IVS5-IVS6 linker (R1740; Hans et al. 1999b), represented by large red circles; the additional valine of a Cav2.1 splice variant converting fast Q-type into slow P-type kinetics (V421; Bourinet et al. 1999), symbolized as a red square; the α-subunit interaction domain (AID) in the I-II loop and additional β-subunit interaction motifs on N- and C-terminal regions of α12.1, shown in green (Pragnell et al. 1994; Walker et al. 1998, 1999); SNARE protein interaction with the Synprint site on the domain II-III linker (Bezprozvanny et al. 1995; Rettig et al. 1996; Zhong et al. 1999), shown in blue; and the calmodulin (CaM)-binding domain (CBD; Lee et al. 1999) in the C-terminus, shown in pink. B, hypothetical scheme of the structural organization of Ca2+ channel segments S1 (white), S5 (green) and S6 (red) and the interaction with intracellular proteins. Analogous to the crystal structure of the KcsA channel (Doyle et al. 1998), the pore-forming S6 segments are arranged as an inverted ‘teepee’ (see also Hering et al. 1998). The loop interactions with intracellular proteins and the location of some crucial inactivation determinants are indicated.
