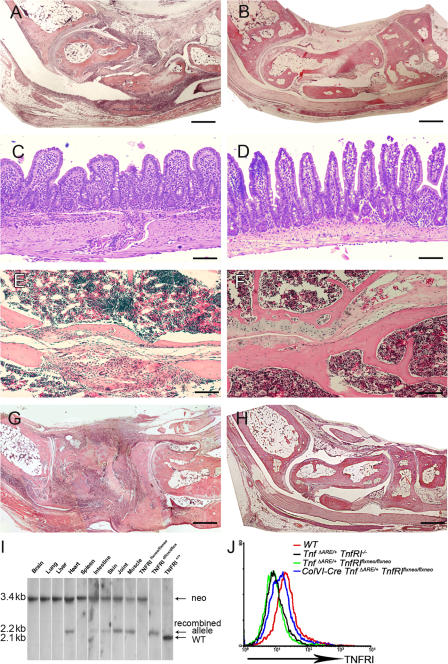
Figure 3.
TNFRI-expressing mesenchymal cells are sufficient targets of pathogenic TNF. (A–D) Histological examination of joint (8-wk-old) and ileal (16-wk-old) sections from ColVI-Cre TnfΔARE/+ TnfRIflxneo/flxneo (A and C) and TnfΔARE/+ TnfRIflxneo/flxneo mice (B and D). (E and F) Histological examination of sacroiliac joint sections from 10-mo-old ColVI-Cre TnfΔARE/+ TnfRIflxneo/flxneo (E) and TnfΔARE/+ TnfRIflxneo/flxneo mice (F). (G and H) Histological examination of joint sections from 7-wk-old ColVI-Cre Tg197 TnfRIflxneo/flxneo (G) and Tg197 TnfRIflxneo/flxneo mice (H). Paraffin sections were stained with hematoxylin and eosin. Bars: (A, B, G, and H) 600 μm; (C–F) 100 μm. (I) Southern blot analysis of BamHI-digested tissue DNA from ColVI-Cre TnfRIflxneo/flxneo mice for detection of the recombined loxP TnfRI allele. (J) Flow cytometric analysis or the detection of TNFRI expression in ColVI-Cre TnfΔARE/+ TnfRIflxneo/flxneo SFs derived from the cultures of individual mice. White lines indicate that intervening lanes have been spliced out.