Abstract
RNA-dependent RNA polymerase 6 (RDR6) catalyses dsRNA synthesis for post-transcriptional gene silencing (PTGS)-associated amplification and the generation of endogeneous siRNAs involved in developmental determinations or stress responses. The functional importance of RDR6 in PTGS led us to examine its connection to the cellular regulatory network by analyzing the hormonal responses of RDR6 gene expression in a cultured cell system. Delivery of dsRNA, prepared in vitro, into cultured rice (Oryza sativa cv. Japonica Dongjin) cells successfully silenced the target isocitrate lyase (ICL) transcripts. Silencing was transient in the absence of abscisic acid (ABA), while it became persistent in the presence of ABA in growth medium. A transcription assay of the OsRDR6 promoter showed that it was positively regulated by ABA. OsRDR6-dependent siRNA(ICL) generation was also significantly up-regulated by ABA. The results showed that, among the five rice OsRDR isogenes, only OsRDR6 was responsible for the observed ABA-mediated amplification and silencing of ICL transcripts. We propose that ABA modulates PTGS through the transcriptional control of the OsRDR6 gene.
INTRODUCTION
Post-transcriptional gene silencing (PTGS) involves degradation or translational repression of target RNA which binds small interfering (si)-RNA or microRNA by Watson–Crick base pairing. While microRNA is derived from genome-encoded precursor RNA with a stem-loop structure, siRNA is from dsRNA trigger, which can be formed either intracellularly or delivered exogeneously. dsRNA is cleaved into siRNAs of 20–25 nt in length by Dicer, a RNAaseIII-like enzyme (1). Down-regulation of target RNA by siRNA occurs within the RNA-induced silencing complex (RISC) containing Argonaute ribonucleoproteins (2). Primary siRNA and dsRNA PTGS trigger often undergo amplification processes through repeated polymerization to synthesize secondary dsRNA by RDR6 and subsequent dicing into secondary siRNAs by Dicer (3). Amplification involves a transitivity mechanism in which the secondary siRNAs being produced move across the original siRNA-target mRNA binding region to either the upstream or downstream direction in plants (4,5). The level of secondary siRNAs may be an important criterion for effective silencing or for cell-to-cell movement of siRNAs (4,6). It has recently been suggested that the fate of target RNA, either used for cleavage or for polymerization, depends on the kinds of Argonaute proteins involved in the guidance of siRNA and target RNA into the RISC (2,7).
RNA-dependent RNA polymerase (RDR) is known to be responsible for the synthesis of dsRNA on ssRNA substrates in either a primer-dependent or primer-independent manner. Such synthesis has been reported in C. elegance, Dictyostelium and plants, but not in Drosophila and mammals (8). There are six RDR isogenes in Arabidopsis—AtRDR1, AtRDR2, AtRDR3a, AtRDR3b, AtRDR3c and AtRDR6, while rice has five homologues—OsRDR1, OsRDR2, OsRDR3a, OsRDR3b and OsRDR6 (8,9). Among these, AtRDR2 is known to participate in transcriptional gene silencing by forming dsRNA from which chromatin-targeting siRNAs are produced (10,11), while AtRDR6 participates in the synthesis of dsRNA from various RNA substrates that are derived from transgene, viral replication, convergent transcription or microRNA-cleaved mRNA (8). It is also suggested that RDR1 plays a role in resistance to herbivore attack in tobacco (12). No function has been assigned for the other RDR isozymes. Over-expression or multi-copies of transgenes tend to generate aberrant RNAs that often contain truncated RNA structures lacking a 3′-polyA tail and/or 5′-cap (13,14). These aberrant RNAs are reported to preferentially recruit RDR6 for dsRNA synthesis. RDR6 prefers to initiate polymerization at the 3′-end in the absence of primer in plants (15–17), although both primed- and unprimed-polymerizations have been demonstrated in vitro. Consistent with these notions, endogeneous mature mRNAs have not been demonstrated to be a target for RDR6-mediated amplification in plants (5).
RDR6 also plays an important role in the control of development progresses or stress resistance by synthesizing dsRNA, which are diced into endogeneous siRNAs such as trans-acting (tasiRNA) (18,19) and natural siRNAs (nat-siRNA) (20), respectively. tasiRNA is derived from the non-coding TAS transcript, which is targeted by a specific microRNA. MicroRNA390 is known to cleave the TAS3 transcript, and the cleavage product is stabilized and polymerized into dsRNA through the activity of RDR6 and Suppressor of Gene Silencing3 (SGS3) (18,19). DCL4, a Dicer-like ribonuclease, dices the dsRNA into tasiRNAs in a phased manner, and these tasiRNAs bind target mRNA for cleavage (21,22). It has also been shown that, as exemplified in Arabidopsis TAS3 RNA, microRNA390-cleaved RNA is a preferred target for RDR6 (23). siRNA–AGO ribonucleoprotein complex recruits RDR6 to initiate primer-dependent polymerization on the microRNA390-cleaved TAS3 RNA substrate in Arabidopsis. One of the tasiRNA targets is an auxin responsive factor3 (ARF3, also called ETTIN) transcript, which is a mediator of auxin signaling involved in cellular differentiation, such as leaf polarity (15,24,25). Because RDR6 is a critical component in the control of developmental processes, it seems logical that its expressional regulation is linked to an internal signaling network. This study demonstrates that ABA up-regulates transcription of the rice OsRDR6 gene, indicating that OsRDR6 gene expression is a target of hormonal control.
MATERIALS AND METHODS
Maintenance of rice suspension cultures and plants
Rice cell culture originated from the ovules (germinal vesicles) of Oryza sativa Japonica. cv. Dongjin, and it was maintained at 25°C with constant agitation in darkness by weekly subculturing. The culture medium contained basal N6 medium components (26) and vitamins, 0.2 mg/l of kinetin, 2 mg/l of 2,4-D and 3% sucrose (pH 5.8). For the acetate culture, sucrose was replaced with 10 mM potassium acetate (pH 5.8).
Formation of dsRNA/cationic peptide complex and its delivery
About 100 ng of POA (polyarginine 12-mer) (synthesized by Peptron, Inc. Korea) were mixed with 100 ng of dsRNA to form a delivery-effective complex in which dsRNA is encased within the POA peptide shell. To deliver less amounts of dsRNA, amounts of POA were accordingly lowered. This complex was confirmed by agarose gel electrophoresis in which dsRNA was not stained with ethidium bromide. The complex was added to 0.5 g of fresh cells and was incubated for 1 h at 25°C in a total volume of 300 μl.
RT-PCR
Total RNA was prepared from the samples using TRI reagent (Molecular Probes Diagnostics, USA) according to the manufacturer's instructions. The first-strand cDNA was synthesized from 2 μg of total RNA in a volume of 20 μl using a evertAid_First Strand cDNA Synthesis Kit (Fermentas, Lithuania). One microliter of the first-strand solution was used for the PCR. The annealing temperature was 55°C for OsRDR6 and ICL, 63°C for OsRDR3a, and 50°C for actin, and 30 cycles of PCR were performed. The following gene-specific primers were used to detect the gene expression level; ICL-FP (5′-CAGCTCAAGACCTTCTCTGA-3′) and ICL-RP (5′-CTCTGGATCCTCTCCACGTA-3′); RDR6-FP (5′-TCACTTAGCTGAGCTAGCAG-3′) and RDR6-RP (5′-TCGGAGATGTATGCTGCAAG-3′); OsRDR3a-FP (5′-TACGGCAGCTGATAGCTGGT-3′) and OsRDR3a-RP (5′-CTGGTAGATCACGCAGGCTT-3′). The rice actin gene (forward, 5′-TCCATCTTGGCATCTCTCAG-3′; reverse, 5′-GTACCCGCATCAGGCATCTG-3′) was used as an internal control.
Detection of siRNA by RNase protection assay
In order to enrich small size-RNA, 50 μg total RNA was mixed with PEG8000 (5%) and NaCl (0.5 M), then incubated on ice for 30 min and centrifuged at 13 000g for 15 min. Small size-RNA was enriched in the supernatant. By adding 10 μg of yeast tRNA and three volumes of EtOH to the supernatant and incubating for 30 min at –70°C, small size-RNA was precipitated and used for the RNase protection assay. The small size-RNA was hybridized to α-32P-UTP-labeled sense RNA (internal region of ICL) overnight at 45°C, and the unhybridized ssRNA portions were digested by RNase A. After the debris was washed out, the hybridized fraction was precipitated with 1 μl of yeast tRNA (10 mg/ml) and three volumes of EtOH. The pellet was dissolved in 10 μl of formaldehyde–RNA dye and loaded into a 15% denaturing PAGE gel, which was exposed to X-ray film.
Transcription assay
Arabidopsis protoplasts were isolated from Arabidopsis 3-week-old seedlings. The whole plant was soaked in 25 ml enzyme solution (1% cellulase R-10, 0.25% macerozyme R-10, 400 mM Mannitol, 8 mM CeCls, 5 mM Mes with pH 5.6 KOH) at 22–25°C for 2–4 h. The protoplasts were carefully filtered with 100 mm mesh to prevent damage and mixed with a final volume of 25 ml W5 solution (154 mM NaCl, 125 mM CaCl2, 5 mM KCl, 5 mM glucose, 1.5 mM Mes with pH 5.6 KOH). Following centrifugation at 40g for 5 min at 25°C, the supernatant was removed by pipette and the remaining pellet was suspended in W5 solution. After checking the protoplasts under an optical microscope, the mixture was gently loaded on top of a 21% sucrose solution and enriched by centrifugation at 40g for 5 min at 25°C. The enriched protoplasts were removed from the middle of the mixture with a pipet, resuspended in 20 ml W5 solution, and stored for 2–4 h at 4°C. The enriched pellet was suspended with MaMg solution (400 mM mannitol, 15 mM MgCl2, 5 mM MES with pH 5.6). Next, plasmid DNA was added, and the mixture was immediately suspended in PEG solution (40% PEG4000, 200 mM Mannitol, 100 mM CaCl2). Then, after flow-through of the same volume of MaMg solution, the mixture was incubated for 30 min at 25°C. The transformed mixture was washed with W5 solution and centrifuged at 40g for 5 min at 25°C, after which the pellet was resuspended in 2 ml of W5 solution.
RDR activity assay
Harvested rice suspension cells were ground by a mortar and pestle in 0.7 volumes of grinding buffer (110 mM KOAc, 20 mM HEPES–KOH at pH 7.3, 1 mM MgOAc, 1 mM DTT, 3 mM EGTA, 2 mM CaCl2). After centrifugation of the homogenate, the supernatant was used for the RDR activity assay. For primer-independent polymerization, the reaction was initiated by adding 50 μg of each cell extract to a reaction mixture containing 5 μg of sense-stranded template RNA (ICL), 2 μl polymerization buffer (50 mM HEPES–KOH at pH 7.8, 20 mM NH4OAc, 5 mM MgCl2, 0.1 mM EDTA, 0.1% Triton X-100), 0.5 mM ATP, 100 μM CTP, 100 μM GTP, 20 μM UTP, 25 μCi α-32P-UTP and 0.2 U/μl RNsin (Ambion) in a total volume of 20 μl. After incubating the mixtures at 25°C for 3 h, the reaction was stopped by vortexing with an equal volume of phenol/chloroform/isoamylalcohol. After centrifugation, the supernatant was precipitated with 1 μl of yeast tRNA (10 mg/ml) and three volumes of EtOH. The pellet was dissolved in 10 μl of formaldehyde–RNA dye and loaded into an 8% denaturing PAGE gel. For siRNA-dependent activity, template RNA, which was annealed to α-32P-UTP labeled siRNA (corresponding to the region 150 bp from the 5′-end of the template) at room temperature for 10 min, was used along with the other reaction components (without α-32P-UTP) to initiate the reaction.
Primer extension
Primer extension analysis was carried out as described previously (27), with some modifications. Total RNA (10 μg) was used for annealing to the 5′-end-labeled primer 1 (5′-TGTCCATGCCCAAACTCCAA-3′) and 5′-end-labeled primer 2 (5′-AAAGCAGCACGGCCATGAAG-3′) at 35°C for 16 h and for subsequent extension using SuperScript II (Invitrogen, CA) at 42°C for 2 h. Samples were separated by 6% denaturing PAGE gel and autoradiographed.
RESULTS AND DISCUSSION
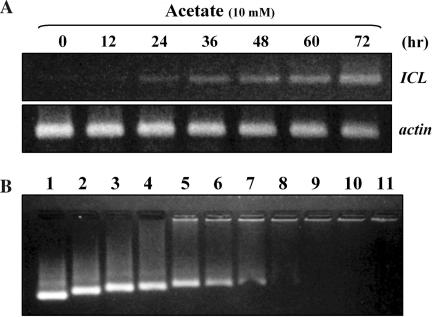
To establish an experimental PTGS system in cultured rice cells, we attempted to silence a gene for isocitrate lyase (ICL), which, as a glyoxylate cycle enzyme, is induced by acetate (Figure 1A). Plant cells are known to operate a glyoxylate cycle to circumvent carbon loss during the citric acid cycle in the presence of acetate as a sole carbon source (28). Acetate-induced ICL mRNA was subjected to PTGS by delivering dsRNA covering the 0.5 kb ICL coding region, which is synthesized in vitro (hereafter designated as dsICL). dsICL was delivered into cells with a polyarginine oligopeptide (POA) carrier, which protects nucleic acids within its shell as a result of positive charges on its surface (29,30). As shown in lane 8 and thereafter in Figure 1B, dsICL appeared to be protected by the POA shell and ready for delivery, as evidenced by negligible ethidium bromide staining in the mobility retardation assay. The ratio of POA/dsICL applied in lane 8 was used for the rest of the studies.
Figure 1.
Formation of polyarginine(POA)/dsICL complex for silencing of ICL. (A) 10 mM acetate in medium gradually up-regulated the level of ICL transcripts during the 72 h period examined. (B) Formation of the dsRNA(ICL)/POA complex. To evaluate optimum complex formation, various concentrations of POA were mixed with a fixed concentration of dsRNA (lane 1, 100 ng of dsRNA only; lanes 2–11, 12.5, 25, 37.5, 50, 62.5, 75, 87.5, 100, 112.5 and 125 ng of POA mixed with 100 ng of dsRNA, respectively). As the POA concentration increased, dsRNA mobility was gradually retarded. The POA concentration in lane 8 was chosen as the most effective delivery-complex.
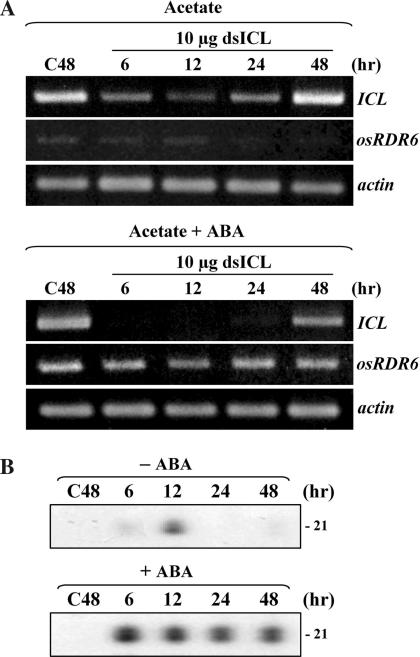
ICL mRNA was silenced when 10 μg of dsICL was delivered into cells growing in acetate (Figure 2A). The ICL mRNA level was down-regulated transiently around 12 h following dsICL delivery. After 48 h, ICL mRNA appeared to recover to its normal level, indicating that silencing was not persistent. The appearance of siRNA was monitored to investigate the progress of PTGS (Figure 2B). Enriched siRNAs were hybridized to α-32P-UTP-labeled sense RNA corresponding to the region used for dsICL preparation, and siRNAs were detected after enzymatic degradation of the unhybridized sense RNA portion. In agreement with the transient decrease of ICL mRNA, siRNA appeared transiently at 12 h (Figure 2B). The OsRDR6 transcript level was down-regulated in the presence of acetate (Figure 3A), strongly suggesting that RDR6-mediated amplification of dsRNAs and siRNAs did not occur in cells grown in acetate. It is very likely that primary siRNAs, derived from the delivered dsICL trigger, were used to target ICL mRNA for cleavage to complete the single round of silencing in the absence of activity of OsRDR6. This PTGS system designed for silencing of ICL transcript was used to investigate expressional control of OsRDR6 and the accompanying PTGS.
Figure 2.
PTGS of ICL transcripts and generation of siRNA. (A) Silencing of ICL transcripts in the absence (upper) and presence (bottom) of ABA. Following delivery of 10 μg of dsICL into cells pre-cultured for 48 h in acetate, the levels of ICL transcripts were monitored by RT–PCR. (B) Detection of siRNA in cells grown in the absence (upper) and presence (bottom) of ABA. Following the delivery of dsICL, appearance of siRNA was monitored. In dsICL-free control cells, siRNA did not appear at 48 h following the transfer into ABA-containing medium (C48).
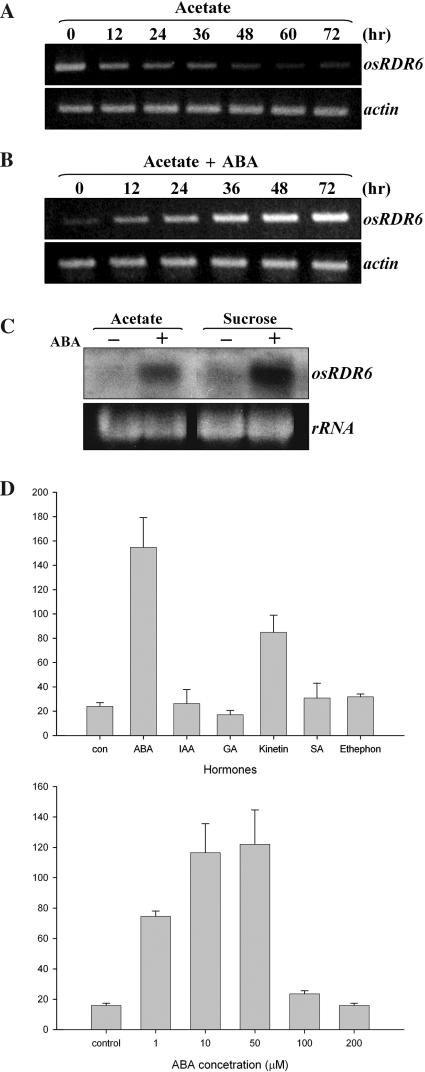
Figure 3.
ABA regulation of OsRDR6 gene expression. RT–PCR analyses of changes of OsRDR6 transcript level following the transfer into acetate medium without ABA (A) and with ABA (B). (C) Northern analyses of ABA up-regulation of OsRDR6 gene expression in cells grown in acetate (10 mM) or sucrose (3%). (D) Transcription assays of OsRDR6 promoter (1.2 kb). A promoter:luciferase construct was transfected into Arabidopsis protoplasts, and they were incubated with different plant hormones (upper), and with different concentrations of ABA (bottom). Variations of the three independent assays are shown.
Initially, the presence of an ABA-responsive DNA cis-element (ACGTG) at the –680 region of OsRDR6 promoter prompted us to examine whether ABA initiated amplification to allow persistent PTGS. RT–PCR analyses showed that the OsRDR6 transcript level was up-regulated in the presence of ABA (Figure 3B). Northern analyses also showed that ABA up-regulated OsRDR6 expression in cells grown either in acetate or sucrose (Figure 3C), indicating that ABA induction was not limited to cells grown in acetate. In line with this observation, the silencing of ICL transcripts was significantly enhanced in the presence of ABA in cells grown in acetate (Figure 2A). It was shown that ICL transcripts were not detected up to 24 h, although they appeared again, at a lower level, at 48 h. siRNAs were detected throughout the examination (Figure 2B), indicating that ABA induced efficient PTGS of ICL transcripts through the persistent generation of siRNA. Re-appearance of the ICL transcript at 48 h (Figure 2A) was likely because PTGS of ICL mRNA was less effective at 48 h. Sufficient amounts of target mRNAs are probably an important factor in the continuous activity of OsRDR6. If secondary siRNAs are defined as ‘siRNAs synthesized from RDR6-generated dsRNA’, the siRNAs produced in the presence of ABA should be considered secondary siRNAs. Although the nature of these siRNAs was not analyzed further, we conclude that these secondary siRNAs were derived from secondary dsRNAs synthesized by ABA-induced activity of OsRDR6. To further confirm ABA control, transcription assays were performed to examine the promoter activity of the OsRDR6 gene. A plasmid vector carrying the 1.2 kb region of the promoter region, which is fused to the luciferase gene, was transfected into Arabidopsis protoplasts to monitor transcriptional activity (Figure 3D). The results showed that OsRDR6 gene was transcriptionally up-regulated by ABA. The optimum concentration of ABA was about 50 μM. Of the other hormones, only kinetin appeared to be somewhat effective.
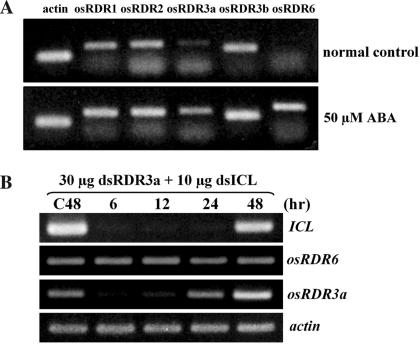
To investigate whether the other OsRDR isogenes, in addition to OsRDR6, are also involved in the amplification of secondary dsICL and siRNAs, the rice RDR homologues were examined for their responses to ABA (Figure 4A). Among the five rice OsRDR homologues, levels of OsRDR6 and OsRDR3a transcripts were significantly up-regulated by ABA, suggesting that OsRDR3a may also participate in amplification in the presence of ABA. To examine the participation of OsRDR3a in PTGS of ICL transcripts, dsRNA spanning the OsRDR3a coding region (dsOsRDR3a) was delivered to silence OsRDR3a transcripts (Figure 4B). Delivered dsOsRDR3a silenced OsRDR3 transcripts efficiently, without silencing OsRDR6 transcripts. OsRDR6-mediated silencing of ICL transcripts in the presence of ABA appeared not to be affected by the silencing of OsRDR3a transcripts. These results strongly suggested that ABA-induced amplification was exclusively due to the activity of OsRDR6. Although OsRDR3a expression was responsive to ABA, its does not appear to participate in the amplification of PTGS. The function of OsRDR3a is yet to be characterized.
Figure 4.
ABA responses of rice RDR homologues. (A) RT–PCR analysis of ABA responsiveness of rice RDR homologues. (B) Silencing of OsRDR3a transcripts by delivering dsOsRDR3a, together with dsICL, into cells growing in acetate in the presence of ABA. Following the delivery of dsOsRDR3a, levels of ICL, OsRDR3a and OsRDR6 transcripts were evaluated by RT–PCR.
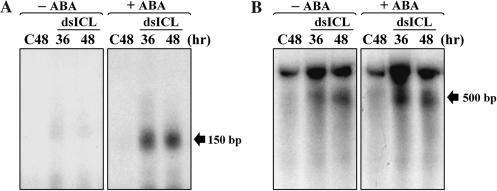
To further characterize ABA up-regulation of OsRDR6 expression, we analyzed RNA polymerization activity (Figure 5). With cell extracts prepared from acetate-grown cells, polymerization of an antisense cRNA strand on the ICL internal sense RNA template (0.5 kb) was examined either with siRNA(ICL) as a primer or without primer. Primer-dependent polymerization was initiated by adding the annealed siRNA(α-32P-labeled)/sense RNA template complex to the extracts, with an expected product of 150 bp (Figure 5A). Primer-dependent activity was clearly induced in cells treated with dsICL and ABA, while it was negligible in cells treated only with dsICL without ABA or in cells treated only with ABA without dsICL. Primer-independent polymerization was similarly up-regulated by ABA in these cells, except that it was also active in cells treated only with dsICL without ABA (Figure 5B). Primer-independent polymerization operated predominantly in the absence of ABA, suggesting that it constitutively operates in the presence of dsICL regardless of the presence of ABA. Taken together, we conclude that ABA activated OsRDR6-mediated polymerization following the transcriptional up-regulation of the OsRDR6 gene.
Figure 5.
Characterization of RNA polymerization, performed with cell extracts in the presence (A) and absence (B) of siRNA primers. Sense-stranded RNA corresponding to the internal ICL region (0.5 kb) was used as a template for polymerization in the polymerization mixture containing α-32P-UTP. In the presence of ABA (right columns), both primed- and unprimed-polymerization appeared to be induced (see 150 and 500 bp expected polymerization products, respectively), while only unprimed polymerization operated in dsICL-delivered cells in the absence of ABA (B, left).
Although both primer-dependent and primer-independent activities were active in these cell extracts, the actual mode of polymerization in vivo is not understood. Primer-independent polymerization is known to be predominant in plants, as aberrant RNAs are usually used as substrates for RDR6 (16). Mature endogeneous mRNAs have never been demonstrated as a substrate for amplification. It is probable that, as an aberrant RNA, delivered dsICL was a favorable substrate for OsRDR6-mediated polymerization to amplify the secondary dsRNAs and secondary siRNAs to degrade large amounts of acetate-inducible ICL mRNA. It remains to be characterized if ICL mRNA is used for amplification by primer-dependent polymerization in the presence of ABA.
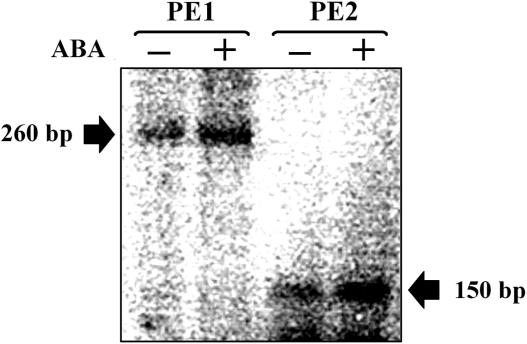
Knowing that RDR6 is also an important component in the generation of endogeneous siRNAs, such as tasiRNAs and nat-siRNAs in Arabidopsis, we wanted to see if this phenomenon also operated in cultured rice cells in relation to ABA. To examine if ABA is also involved in the up-regulation of OsRDR6 expression to enhance the efficiency of tasiRNA generation, we chose a pathway in which microRNA390-cleaved TAS3 precursor is cleaved into tasiRNAs. RDR6-mediated tasiRNA synthesis, in conjunction with DCL4 and AGO1/7, is responsible for several aspects of plant development in Arabidopsis (31). A group of these tasiRNAs is known to target ARF3 mRNA for cleavage in Arabidopsis (designated tasiRNA-ARF). However, we were not able to detect tasiRNA-ARF in cultured rice cells treated with ABA, probably because the level of tasiRNA-ARF was too low to detect in these cultured cells even in the presence of ABA. On the assumption that tasiRNA-ARF synthesis was up-regulated by ABA, levels of the cleavage products of ARF3 mRNA were alternatively analyzed for the ABA response, since cleavage of ARF3 mRNA is derived from its binding to tasiRNA-ARF. The suggested cleavage site for tasiRNA-ARF within the ARF3 mRNA in Arabidopsis was also confirmed in rice cells (Figure 6), indicating that this endogeneous pathway is conserved among these plants. Primer extension assays were performed to detect the primer-extended cleavage fragments, which spans from the primer-binding site to the expected cleavage site (Figure 6). Two different primers were used to produce the expected sizes of 260 bp (PE1) and 150 bp (PE2). The levels of the two predicted cleavage fragments were significantly higher in ABA-treated cells, strongly suggesting that ABA-induction of the OsRDR6 gene was responsible for enhanced synthesis of tasiRNA-ARF and cleavage of ARF3 mRNA.
Figure 6.
ABA regulation of tasiRNA-ARF generation. The tasiRNA-ARF level was measured by monitoring cleavage of ARF3 mRNA by primer extension (PE) assays. Two primers were designed to extend to the predicted cleavage sites and generate products of 260 bp (PE1) and 150 bp (PE2) (arrows).
This study clearly demonstrates that transcription of the OsRDR6 gene is positively regulated by ABA. Cleavage of target transcripts by siRNAs, which are derived from OsRDR6-generated dsRNA, was accordingly up-regulated by ABA. This study provides a mechanistic model in which PTGS duration is extended by up-regulating RDR6 gene expression. QDE-1, a RDR6 equivalent in Neurospora crassa, is a rate-limiting factor for QDE-1-mediated PTGS, and the efficiency and persistence of PTGS depends on the amount of cellular QDE-1 protein (32). Since ABA signaling is widely interconnected with other hormones and internal regulatory networks, the results demonstrated in this study suggest that PTGS is closely associated with a cellular signaling network at least via OsRDR6. Although other PTGS components were not investigated, transcriptional control of the OsRDR6 gene appears to be a major target for hormonal control. Hormones may participate in gene silencing by regulating the HYL1 protein, which, as a dsRNA-binding protein, is known to affect accumulation of microRNA (33). Mutation in the hyl1 gene disrupts plant sensitivity to ABA, auxin and cytokinin, implying that the biogenesis of microRNA is linked to hormonal regulation. ABA is known to control gene expression of several RNA binding proteins, suggesting that ABA is involved in many aspects of RNA processing, such as pre-mRNA splicing, 5-capping, 3′-polyA tailing, or export from the nucleus (34). It was also shown that ABA directly binds a nuclear RNA binding protein FCA to control autonomous floral pathway (35). As this study indicates, ABA is also involved in determining the cellular level of secondary siRNAs via transcriptional up-regulation of OsRDR6 gene. We conclude that PTGS involving RDR6-mediated RNA polymerization is closely associated with a hormonal signaling network.
ACKNOWLEDGEMENTS
Funding for this work was provided by The Ministry of Science & Technology and Korea Science & Engineering Foundation, Korea. This work was also partly supported by a SungKyun Academic Research Grant, Sungkyunkwan University. J.H.Y. and S.J.H. were recipients of a Graduate Fellowship awarded by the Brain Korea 21 Program of the Ministry of Education & Human Resources Development, Korea. Funding to pay the Open Access publication charges for this article was provided by Sungkyunkwan University.
Conflict of interest statement. None declared.
REFERENCES
- 1.Schauer SE, Jacobsen SE, Meinke DW, Ray A. DICER-LIKE1: blind men and elephants in Arabidopsis development. Trends Plant Sci. 2002;7:487–491. doi: 10.1016/s1360-1385(02)02355-5. [DOI] [PubMed] [Google Scholar]
- 2.Baumberger N, Baulcombe DC. Arabidopsis ARGONAUTE1 is an RNA slicer that selectively recruits microRNAs and short interfering RNAs. Proc. Natl. Acad. Sci. USA. 2005;102:11928–11933. doi: 10.1073/pnas.0505461102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sijen T, Fleenor J, Simmer F, Thijssen KL, Parrish S, Timmons L, Plasterk RH, Fire A. On the role of RNA amplification in dsRNA-triggered gene silencing. Cell. 2001;107:465–476. doi: 10.1016/s0092-8674(01)00576-1. [DOI] [PubMed] [Google Scholar]
- 4.Garcia-Perez RD, Houdt HV, Depicker A. Spreading of post-transcriptional gene silencing along the target gene promotes systemic silencing. Plant J. 2004;38:594–602. doi: 10.1111/j.1365-313X.2004.02067.x. [DOI] [PubMed] [Google Scholar]
- 5.Vaistij FE, Jones L, Baulcombe DC. Spreading of RNA targeting and DNA methylation in RNA silencing requires transcription of the target gene and a putative RNA-dependent RNA polymerase. Plant Cell. 2002;14:857–867. doi: 10.1105/tpc.010480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Himber C, Dunoyer P, Moissiard G, Ritzenthaler C, Voinnet O. Transitivity-dependent and -independent cell-to-cell movement of RNA silencing. EMBO J. 2003;22:4523–4533. doi: 10.1093/emboj/cdg431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20:515–524. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- 8.Wassenegger M, Krczal G. Nomenclature and functions of RNA-directed RNA polymerases. Trends Plant Sci. 2006;11:142–151. doi: 10.1016/j.tplants.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 9.Makeyev EV, Bamford DH. Cellular RNA-dependent RNA polymerase involved in posttranscriptional gene silencing has two distinct activity modes. Mol. Cell. 2002;10:1417–1427. doi: 10.1016/s1097-2765(02)00780-3. [DOI] [PubMed] [Google Scholar]
- 10.Chan SW, Zilberman D, Xie Z, Johansen LK, Carrington JC, Jacobsen SE. RNA silencing genes control de novo DNA methylation. Science. 2004;303:1336. doi: 10.1126/science.1095989. [DOI] [PubMed] [Google Scholar]
- 11.Xie Z, Johansen LK, Gustafson AM, Kasschau KD, Lellis AD, Ziberman D, Jacobsen SE, Carrington JC. Genetic and functional diversification of small RNA pathways in plants. PLos Biol. 2004;2:E104. doi: 10.1371/journal.pbio.0020104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pandey SP, Baldwin IT. RNA-directed RNA polymerase 1 (RdR1) mediates the resistance of Nicotiana attenuate to herbivore attack in nature. Plant Cell. 2007;50:40–53. doi: 10.1111/j.1365-313X.2007.03030.x. [DOI] [PubMed] [Google Scholar]
- 13.Beclin C, Boutet S, Waterhouse P, Vaucheret H. A branched pathway for transgene-induced RNA silencing in plants. Curr. Biol. 2002;12:684–688. doi: 10.1016/s0960-9822(02)00792-3. [DOI] [PubMed] [Google Scholar]
- 14.Luo Z, Chen Z. Improperly terminated, unpolyadenylated mRNA of sense transgenes is targeted by RDR6-mediated RNA silencing in Arabidopsis. Plant Cell. 2007;19:943–958. doi: 10.1105/tpc.106.045724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Allen E, Xie Z, Gustafson AM, Carrington JC. microRNA-directed phasing during Trans-Acting siRNA biogenesis in plants. Cell. 2005;121:207–221. doi: 10.1016/j.cell.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 16.Petersen BO, Albrechtsen M. Evidence implying only unprimed RdRP activity during transitive gene silencing in plants. Plant Mol. Biol. 2005;58:575–583. doi: 10.1007/s11103-005-7307-4. [DOI] [PubMed] [Google Scholar]
- 17.Tang G, Reinhart BJ, Bartel DP, Zamore PD. A biochemical framework for RNA silencing in plants. Genes Dev. 2003;17:49–63. doi: 10.1101/gad.1048103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peragine A, Yoshikawa M, Wu G, Albrecht HL, Poethig RS. SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev. 2004;18:2368–2379. doi: 10.1101/gad.1231804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vazquez F, Vaucheret H, Rajagopalan R, Lepers C, Gasciolli V, Mallory AC, Hilbert JL, Bartel DP, Crete P. Endogeneous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol. Cell. 2004;16:69–79. doi: 10.1016/j.molcel.2004.09.028. [DOI] [PubMed] [Google Scholar]
- 20.Borsani O, Zhu J, Verslues PE, Sunkar R, Zhu JK. Endogeneous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell. 2005;123:1279–1291. doi: 10.1016/j.cell.2005.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gasciolli, Mallory AC, Bartel DP, Vaucheret H. Partially redundant functions of Arabidopsis DICER-like enzymes and a role for DCL4 in producing trans-acting siRNAs. Curr. Biol. 2005;15:1494–1500. doi: 10.1016/j.cub.2005.07.024. [DOI] [PubMed] [Google Scholar]
- 22.Xie Z, Allen E, Wilken A, Carrington JC. Dicer-like 4 functions in trans-acting small interfering RNA biogenesis and vegetative phase change in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA. 2005;102:12984–12989. doi: 10.1073/pnas.0506426102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Axtell MJ, Jan C, Rajagopalan R, Bartel DP. A two-hit trigger for siRNA biogenesis in plants. Cell. 2006;127:565–577. doi: 10.1016/j.cell.2006.09.032. [DOI] [PubMed] [Google Scholar]
- 24.Williams L, Carles CC, Osmont KS, Fletcher JC. A database analysis method identifies an endogeneous trans-acting short-interfering RNA that targets the Arabidopsis ARF2, ARF3, and ARF4 genes. Proc. Natl. Acad. Sci. USA. 2005;102:9703–9708. doi: 10.1073/pnas.0504029102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fahlgren N, Montgomery TA, Howell MD, Allen E, Dvorak SK, Alexander AL, Carrington JC. Regulation of AUXIN RESPONSE FACTOR3 by TAS3 ta-siRNA affects developmental timing and patterning in Arabidopsis. Curr. Biol. 2006;16:939–944. doi: 10.1016/j.cub.2006.03.065. [DOI] [PubMed] [Google Scholar]
- 26.Chu CC, Wang CS, Sun CS, Hus C, Ying KC, Chu CY, Bi FY. Establishment of an efficient medium for anther culture of rice through comparative experiments on the nitrogen sources. Scienta Sinic. 1975;18:659–668. [Google Scholar]
- 27.Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee TK, Lee WS. Diauxic growth in rice suspension cells grown on mixed carbon sources of acetate and glucose. Plant Physiol. 1996;110:465–470. doi: 10.1104/pp.110.2.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schwarze SR, Ho A, Vocero-Akbani A, Dowdy SF. In vivo protein transduction: delivery of a biologically active protein into the mouse. Science. 1999;285:1569–1572. doi: 10.1126/science.285.5433.1569. [DOI] [PubMed] [Google Scholar]
- 30.Unnamalai N, Kang BG, Lee WS. Cationic oligopeptide-mediated delivery of dsRNA for post-transcriptional gene silencing in plant cells. FEBS Lett. 2004;566:307–310. doi: 10.1016/j.febslet.2004.04.018. [DOI] [PubMed] [Google Scholar]
- 31.Hunter C, Willmann MR, Wu G, Yoshikawa M, de la Luz Gutierrez-Nava M, Poethig SR. Trans-acting siRNA-mediated repression of ETTIN and ARF4 regulates heteroblasty in Arabidopsis. Development. 2006;133:2973–2981. doi: 10.1242/dev.02491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Forrest EC, Cogoni C, Macino G. The RNA-dependent RNA polymerase, QDE-1, is a rate-limiting factor in post-transcriptional gene silencing in Neurospora crassa. Nucleic Acids Res. 2004;32:2123–2128. doi: 10.1093/nar/gkh530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu C, Fedoroff N. A mutation in the Arabidopsis HYL1 gene encoding a dsRNA binding protein affects responses to abscisic acid, auxin, and cytokinin. Plant Cell. 2000;12:2351–2366. doi: 10.1105/tpc.12.12.2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuhn JM, Schroeder JI. Impacts of altered RNA metabolism on abscisic acid signaling. Curr. Opin. Plant Biol. 2003;6:463–469. doi: 10.1016/s1369-5266(03)00084-0. [DOI] [PubMed] [Google Scholar]
- 35.Razem FA, El-Kereamy A, Abrams SR, Hill RD. The RNA-binding protein FCA is an abscisic acid receptor. Nature. 2006;439:290–294. doi: 10.1038/nature04373. [DOI] [PubMed] [Google Scholar]