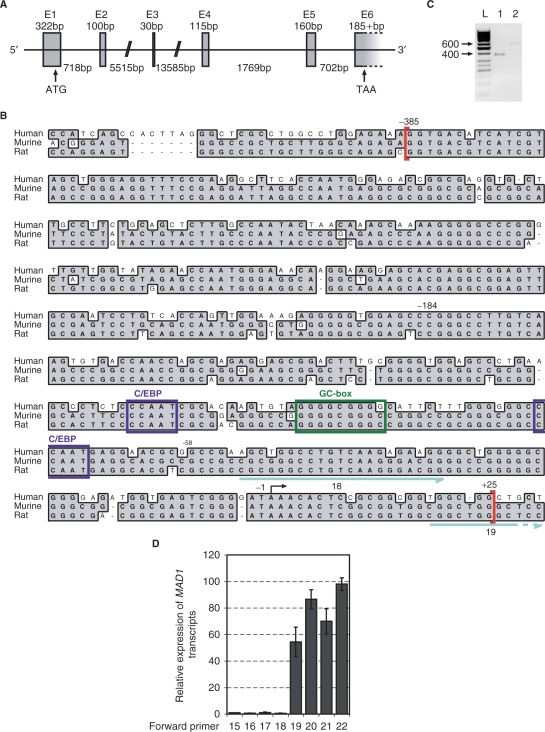
Figure 2.
Identification of the putative promoter region of MAD1. (A) The genomic organization of human MAD1 was deduced from comparing the cDNA to genomic clone HTGS Acc. No. AC019206. The ATG start codon is indicated in exon 1. E, exon sequences. (B) Upstream of the most 5′ cDNA sequences of human, rat and mouse MAD1, a genomic region of sequences homology is shown. This region encompasses nucleotides –385 to +25 relative to the start of transcription and is indicated with red brackets. The positions of nucleotides –184 and –58 are indicated, which delineate the region that is responsive to G-CSF/G-CSFR-mediated signals and contain two CCAAT- and a GC-box. The start site of transcription that was identified by 5′RACE is indicated by an arrow. The number of CpG dinucleotides within the homology region of the three genomic regions is: 23 out of 410 nt in human, 43 out of 405 in rat and 45 out of 405 in mouse. In addition, two of the primers (18 and 19, blue arrows) are indicated that were used to analyze 5'-RACE products (see panel D). (C) Total RNA was generated from G-CSF-treated U937 cells and reverse transcribed. 5′ RACE was performed and the main products were identified and sequenced. The results of two independent reactions are shown. Sequencing of the 600 bp product revealed the start site of transcription indicated in panel B. (D) The primary 5′-RACE products were amplified with the indicated forward primers and the Sp2 reverse primer by qPCR. The amount of products obtained with primer 15 was set as 1. Mean values and standard deviations of two independent experiments performed in duplicates are displayed.