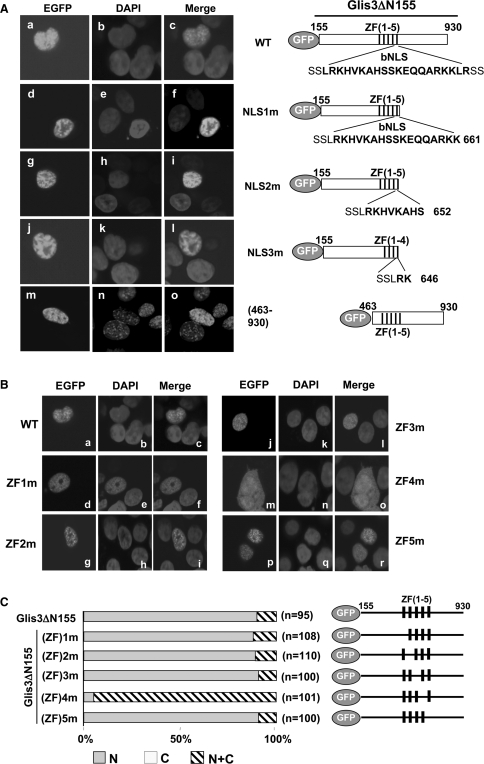
Figure 2.
Nuclear localization of Glis3 does not require the putative bNLS but is dependent on ZF4. HEK293T cells were transfected with the pEGFP-Glis3 plasmid indicated and 30 h later examined by confocal microscopy for EGFP and DAPI fluorescence as described in ‘Materials and Methods’ section. EGFP was equally divided between cytoplasm and nucleus (data not shown). (A) The bNLS is not required for the nuclear localization of Glis3. Subcellular localization of wild-type (WT) EGFP-Glis3 (a–c), EGFP-Glis3-NLS1m (d–f), EGFP-Glis3-NLS2m (g–i), EGFP-Glis3-NLS3m (j–l), EGFP-Glis3(463–930) (m–o). A schematic view of each construct is shown on the right. The putative bNLS sequence is shown in bold. (B) The nuclear localization of Glis3 is dependent on the tetrahedral configuration of ZF4. Subcellular localization of EGFP-Glis3 (a–c), EGFP-Glis3(ZF)1m (d–f), EGFP-Glis3(ZF)2m (g–i), EGFP-Glis3(ZF)3m (j–l), EGFP-Glis3(ZF)4m (m–o) and EGFP-Glis3(ZF)5m (p–r). A schematic view of EGFP-Glis3(ZF)1–5m mutants is shown on the right in (C). The first Cys in each zinc finger motif is mutated to Ala causing loss of the tetrahedral zinc finger configuration as indicated. (C) Quantitation of the subcellular localization of Glis3 and Glis3 mutants. The percentage of cells in which Glis3 is largely restricted to the nucleus (N) or about equally distributed between the nucleus and cytoplasm (N+C) was calculated. ‘n’ indicates the number of Glis3 expressing cells counted.