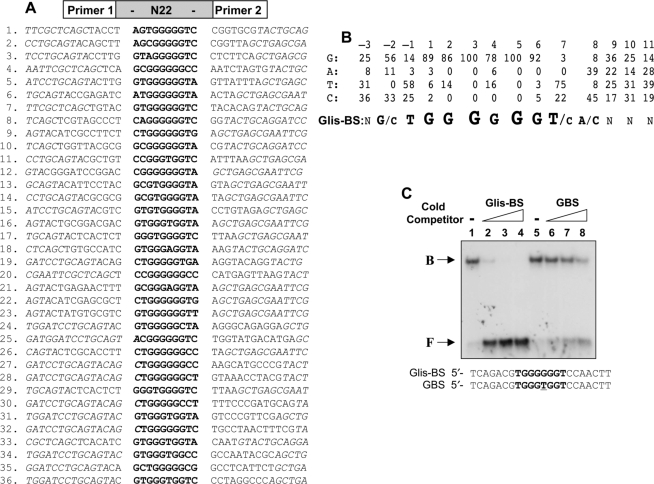
Figure 4.
Identification of the consensus DNA-binding site (Glis-BS). To determine the consensus sequence of the Glis-BS that binds Glis3 with highest affinity, a DNA-binding site selection strategy was followed that was based on a combination of PCR and EMSA as described in ‘Materials and Methods’ section. A mixture of 60 bp oligonucleotides containing 22 random bases (N22) flanked on each side by a 20 bp primer (primer 1 and 2) was used as template. (A) Sequences of the 22 nt long fragments that bind Glis3 with high affinity identified in 36 independent clones. The Glis-BSs are indicated in bold. Italic nucleotides are part of the primer sequence. (B) Calculation of the consensus Glis-BS. Top line: numbers indicate the position of the nucleotide in the Glis-BS. The next four lines indicate the percentages by which G, A, T or C were present at each position. The bottom line shows the calculated consensus sequence of Glis-BS. The font size relates to the frequency by which the nucleotide is found at each position. N indicates no strong requirement for a specific nucleotide. (C) Binding of His6-Glis3(ZFD) (2 μg) to the 32P-labeled consensus Glis-BS (0.5 nM) was examined by EMSA in the absence (lane 1) and presence of a 5-, 25- and 50-fold excess of unlabeled consensus Glis-BS (lanes 2–4) or GBS (lanes 6–8). The sequence of Glis-BS and GBS is shown at the bottom. B, indicates bound Glis-BS or GBS; F, free Glis-BS or GBS.